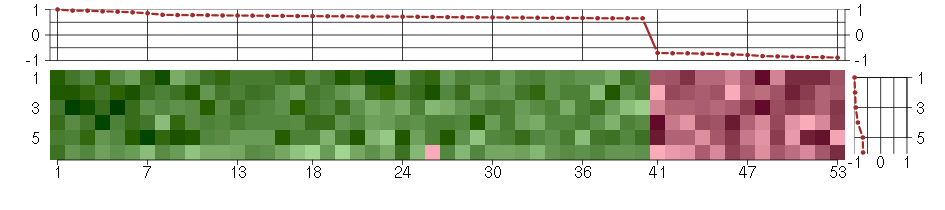

Under-expression is coded with green,
over-expression with red color.



ABLIM1actin binding LIM protein 1 (200965_s_at), score: 0.7 ADCY3adenylate cyclase 3 (209321_s_at), score: 0.93 ATP6V0E1ATPase, H+ transporting, lysosomal 9kDa, V0 subunit e1 (214149_s_at), score: 0.72 BIN1bridging integrator 1 (210201_x_at), score: 0.74 BTBD2BTB (POZ) domain containing 2 (207722_s_at), score: 0.67 C1orf38chromosome 1 open reading frame 38 (210785_s_at), score: 0.66 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (217849_s_at), score: 0.68 CHST2carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2 (203921_at), score: 0.75 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: 0.68 COL10A1collagen, type X, alpha 1 (217428_s_at), score: 0.69 DENND4BDENN/MADD domain containing 4B (202860_at), score: 0.74 DMWDdystrophia myotonica, WD repeat containing (33768_at), score: 0.95 ELL3elongation factor RNA polymerase II-like 3 (219517_at), score: 0.66 EPHA4EPH receptor A4 (206114_at), score: -0.72 FKBPLFK506 binding protein like (219187_at), score: -0.74 FKRPfukutin related protein (219853_at), score: 0.78 FLJ12529pre-mRNA cleavage factor I, 59 kDa subunit (217866_at), score: 0.72 FOXK2forkhead box K2 (203064_s_at), score: 1 GOLPH3golgi phosphoprotein 3 (coat-protein) (217803_at), score: 0.96 HIST1H2BIhistone cluster 1, H2bi (208523_x_at), score: -0.7 HSD17B6hydroxysteroid (17-beta) dehydrogenase 6 homolog (mouse) (37512_at), score: -0.78 HSPA12Aheat shock 70kDa protein 12A (214434_at), score: 0.79 KATNB1katanin p80 (WD repeat containing) subunit B 1 (203162_s_at), score: -0.87 KCNN2potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2 (220116_at), score: -0.73 KIAA0317KIAA0317 (202128_at), score: 0.67 KIAA1305KIAA1305 (220911_s_at), score: 0.75 MAP2K3mitogen-activated protein kinase kinase 3 (207667_s_at), score: 0.69 MAST4microtubule associated serine/threonine kinase family member 4 (40016_g_at), score: 0.66 MC4Rmelanocortin 4 receptor (221467_at), score: -0.85 NEFMneurofilament, medium polypeptide (205113_at), score: -0.87 NUDT11nudix (nucleoside diphosphate linked moiety X)-type motif 11 (219855_at), score: 0.72 NUPL2nucleoporin like 2 (204003_s_at), score: -0.76 PHC1polyhomeotic homolog 1 (Drosophila) (218338_at), score: 0.85 PIGLphosphatidylinositol glycan anchor biosynthesis, class L (213889_at), score: 0.71 POM121POM121 membrane glycoprotein (rat) (212178_s_at), score: 0.71 RAD54L2RAD54-like 2 (S. cerevisiae) (213205_s_at), score: 0.74 REREarginine-glutamic acid dipeptide (RE) repeats (200940_s_at), score: 0.74 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: -0.72 SCAMP4secretory carrier membrane protein 4 (213244_at), score: 0.73 SIRT6sirtuin (silent mating type information regulation 2 homolog) 6 (S. cerevisiae) (219613_s_at), score: 0.89 SMG7Smg-7 homolog, nonsense mediated mRNA decay factor (C. elegans) (201794_s_at), score: 0.78 SNX27sorting nexin family member 27 (221006_s_at), score: 0.77 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: 0.76 TBX3T-box 3 (219682_s_at), score: 0.76 TMEM149transmembrane protein 149 (219690_at), score: -0.89 TNIP1TNFAIP3 interacting protein 1 (207196_s_at), score: 0.66 TOP3Atopoisomerase (DNA) III alpha (214300_s_at), score: -0.83 TSKUtsukushin (218245_at), score: 0.91 TXLNAtaxilin alpha (212300_at), score: 0.68 VPS37Bvacuolar protein sorting 37 homolog B (S. cerevisiae) (221704_s_at), score: 0.7 WDR6WD repeat domain 6 (217734_s_at), score: 0.78 YPEL1yippee-like 1 (Drosophila) (206063_x_at), score: -0.84 ZNF814zinc finger protein 814 (60794_f_at), score: 0.66
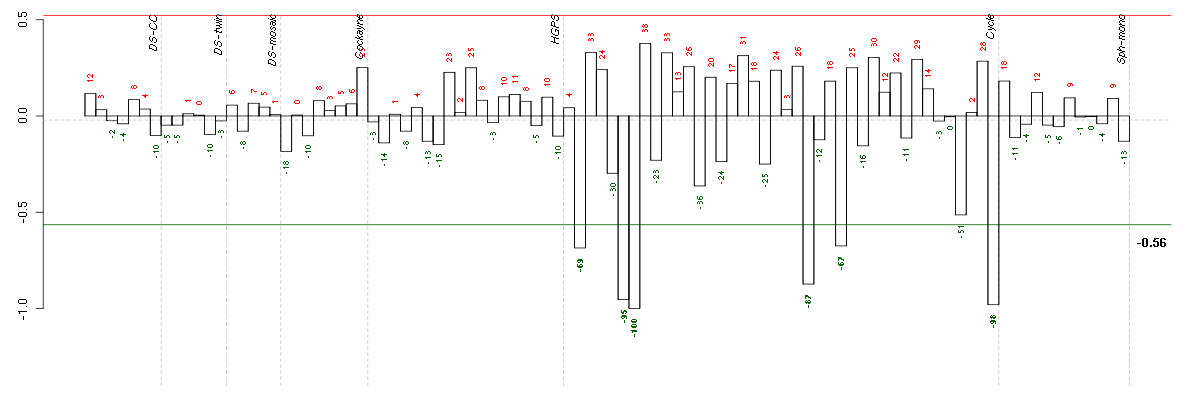
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486151.cel | 26 | 6 | Cycle | hgu133a2 | none | Cycle 1 |