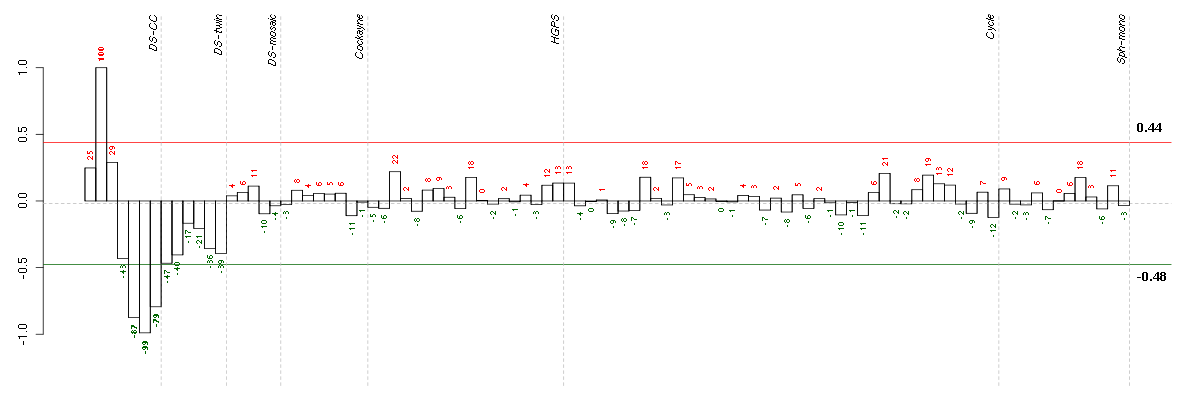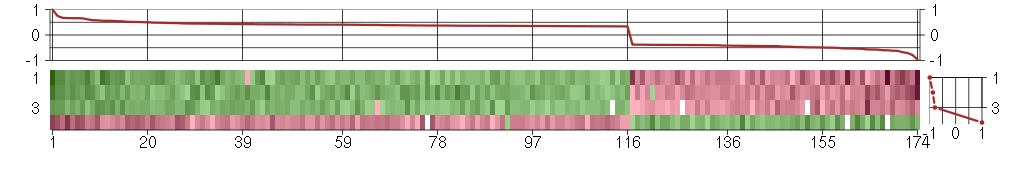

Under-expression is coded with green,
over-expression with red color.

immune system process
Any process involved in the development or functioning of the immune system, an organismal system for calibrated responses to potential internal or invasive threats.
cell adhesion
The attachment of a cell, either to another cell or to an underlying substrate such as the extracellular matrix, via cell adhesion molecules.
immune response
Any immune system process that functions in the calibrated response of an organism to a potential internal or invasive threat.
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cell proliferation
The multiplication or reproduction of cells, resulting in the expansion of a cell population.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
biological adhesion
The attachment of a cell or organism to a substrate or other organism.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
developmental process
A biological process whose specific outcome is the progression of an integrated living unit: an anatomical structure (which may be a subcellular structure, cell, tissue, or organ), or organism over time from an initial condition to a later condition.
organ development
Development of a tissue or tissues that work together to perform a specific function or functions. Development pertains to the process whose specific outcome is the progression of a structure over time, from its formation to the mature structure. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
anatomical structure development
The biological process whose specific outcome is the progression of an anatomical structure from an initial condition to its mature state. This process begins with the formation of the structure and ends with the mature structure, whatever form that may be including its natural destruction. An anatomical structure is any biological entity that occupies space and is distinguished from its surroundings. Anatomical structures can be macroscopic such as a carpel, or microscopic such as an acrosome.
response to stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
all
This term is the most general term possible
cell adhesion
The attachment of a cell, either to another cell or to an underlying substrate such as the extracellular matrix, via cell adhesion molecules.
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
immune response
Any immune system process that functions in the calibrated response of an organism to a potential internal or invasive threat.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
organ development
Development of a tissue or tissues that work together to perform a specific function or functions. Development pertains to the process whose specific outcome is the progression of a structure over time, from its formation to the mature structure. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.

extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
proteinaceous extracellular matrix
A layer consisting mainly of proteins (especially collagen) and glycosaminoglycans (mostly as proteoglycans) that forms a sheet underlying or overlying cells such as endothelial and epithelial cells. The proteins are secreted by cells in the vicinity.
extracellular space
That part of a multicellular organism outside the cells proper, usually taken to be outside the plasma membranes, and occupied by fluid.
extracellular matrix
A structure lying external to one or more cells, which provides structural support for cells or tissues; may be completely external to the cell (as in animals) or be part of the cell (as in plants).
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
all
This term is the most general term possible
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.

protein binding
Interacting selectively with any protein or protein complex (a complex of two or more proteins that may include other nonprotein molecules).
molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
receptor binding
Interacting selectively with one or more specific sites on a receptor molecule, a macromolecule that undergoes combination with a hormone, neurotransmitter, drug or intracellular messenger to initiate a change in cell function.
cytokine activity
Functions to control the survival, growth, differentiation and effector function of tissues and cells.
binding
The selective, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
cytokine binding
Interacting selectively with a cytokine, any of a group of proteins that function to control the survival, growth and differentiation of tissues and cells, and which have autocrine and paracrine activity.
all
This term is the most general term possible
| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 04060 | 4.233e-03 | 2.61 | 11 | 115 | Cytokine-cytokine receptor interaction |
ACSL4acyl-CoA synthetase long-chain family member 4 (202422_s_at), score: 0.41 ADH1Balcohol dehydrogenase 1B (class I), beta polypeptide (209612_s_at), score: 0.66 ADRA2Aadrenergic, alpha-2A-, receptor (209869_at), score: 0.41 AKR1C3aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II) (209160_at), score: 0.51 APODapolipoprotein D (201525_at), score: 0.75 ASAP2ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 (206414_s_at), score: 0.62 ASNSasparagine synthetase (205047_s_at), score: -0.4 ATF5activating transcription factor 5 (204999_s_at), score: 0.47 B4GALT6UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 (206233_at), score: -0.47 BCL3B-cell CLL/lymphoma 3 (204908_s_at), score: 0.35 BNC1basonuclin 1 (206581_at), score: 0.34 BNIP3BCL2/adenovirus E1B 19kDa interacting protein 3 (201848_s_at), score: -0.42 BTN3A2butyrophilin, subfamily 3, member A2 (209846_s_at), score: 0.35 C13orf15chromosome 13 open reading frame 15 (218723_s_at), score: 0.39 C22orf9chromosome 22 open reading frame 9 (217118_s_at), score: 0.34 C2CD2C2 calcium-dependent domain containing 2 (212875_s_at), score: 0.4 CAB39calcium binding protein 39 (217873_at), score: 0.36 CADM1cell adhesion molecule 1 (209031_at), score: -0.48 CCND2cyclin D2 (200953_s_at), score: -0.71 CCRL1chemokine (C-C motif) receptor-like 1 (220351_at), score: 0.66 CD302CD302 molecule (203799_at), score: 0.38 CDC42SE1CDC42 small effector 1 (218157_x_at), score: 0.38 CDH18cadherin 18, type 2 (206280_at), score: -0.57 CEBPZCCAAT/enhancer binding protein (C/EBP), zeta (203341_at), score: 0.42 CHORDC1cysteine and histidine-rich domain (CHORD)-containing 1 (218566_s_at), score: 0.36 CLEC2BC-type lectin domain family 2, member B (209732_at), score: 0.4 CNN3calponin 3, acidic (201445_at), score: -0.39 COL11A1collagen, type XI, alpha 1 (37892_at), score: -0.5 COL14A1collagen, type XIV, alpha 1 (212865_s_at), score: 0.58 CPNE3copine III (202118_s_at), score: 0.37 CREG1cellular repressor of E1A-stimulated genes 1 (201200_at), score: 0.36 CSF1colony stimulating factor 1 (macrophage) (209716_at), score: 0.48 CSRP2cysteine and glycine-rich protein 2 (211126_s_at), score: -0.49 CTSBcathepsin B (213274_s_at), score: 0.5 CTSL1cathepsin L1 (202087_s_at), score: 0.54 CXCL1chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) (204470_at), score: 0.4 CXCL3chemokine (C-X-C motif) ligand 3 (207850_at), score: 0.36 DKK2dickkopf homolog 2 (Xenopus laevis) (219908_at), score: 0.67 DNTTIP2deoxynucleotidyltransferase, terminal, interacting protein 2 (202776_at), score: 0.34 DOCK4dedicator of cytokinesis 4 (205003_at), score: 0.34 DPP4dipeptidyl-peptidase 4 (211478_s_at), score: 0.45 DPTdermatopontin (213068_at), score: 0.35 DSPdesmoplakin (200606_at), score: -0.46 EDIL3EGF-like repeats and discoidin I-like domains 3 (207379_at), score: -0.49 EGFRepidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) (201983_s_at), score: 0.47 EGR1early growth response 1 (201694_s_at), score: -0.39 EIF4EBP1eukaryotic translation initiation factor 4E binding protein 1 (221539_at), score: -0.39 EIF5Aeukaryotic translation initiation factor 5A (201123_s_at), score: -0.54 ELK3ELK3, ETS-domain protein (SRF accessory protein 2) (221773_at), score: 0.37 EML1echinoderm microtubule associated protein like 1 (204797_s_at), score: 0.37 ETS2v-ets erythroblastosis virus E26 oncogene homolog 2 (avian) (201328_at), score: 0.37 F3coagulation factor III (thromboplastin, tissue factor) (204363_at), score: 0.44 FAM38Bfamily with sequence similarity 38, member B (219602_s_at), score: -0.49 FBLN2fibulin 2 (203886_s_at), score: 0.35 FGF13fibroblast growth factor 13 (205110_s_at), score: 0.37 FLGfilaggrin (215704_at), score: -0.96 FMODfibromodulin (202709_at), score: -0.43 FNDC3Afibronectin type III domain containing 3A (202304_at), score: 0.42 FOSv-fos FBJ murine osteosarcoma viral oncogene homolog (209189_at), score: -0.67 FOSL1FOS-like antigen 1 (204420_at), score: 0.41 GLSglutaminase (203159_at), score: -0.43 GOLSYNGolgi-localized protein (218692_at), score: -0.53 GPNMBglycoprotein (transmembrane) nmb (201141_at), score: 0.35 GPRC5BG protein-coupled receptor, family C, group 5, member B (203632_s_at), score: 0.36 HAPLN1hyaluronan and proteoglycan link protein 1 (205523_at), score: -0.54 HK2hexokinase 2 (202934_at), score: 0.44 HMGN1high-mobility group nucleosome binding domain 1 (200944_s_at), score: -0.45 HOXC10homeobox C10 (218959_at), score: 0.5 HOXC8homeobox C8 (221350_at), score: -0.43 HPCAL1hippocalcin-like 1 (205462_s_at), score: 0.41 HSD17B11hydroxysteroid (17-beta) dehydrogenase 11 (217989_at), score: 0.36 HSPB7heat shock 27kDa protein family, member 7 (cardiovascular) (218934_s_at), score: 0.39 HTR2A5-hydroxytryptamine (serotonin) receptor 2A (207135_at), score: 0.34 IER5immediate early response 5 (218611_at), score: 0.35 IFITM1interferon induced transmembrane protein 1 (9-27) (214022_s_at), score: 0.36 IGFBP7insulin-like growth factor binding protein 7 (201162_at), score: -0.42 IL15RAinterleukin 15 receptor, alpha (207375_s_at), score: 0.42 IL1R1interleukin 1 receptor, type I (202948_at), score: 0.35 IL1RL1interleukin 1 receptor-like 1 (207526_s_at), score: 1 IL33interleukin 33 (209821_at), score: 0.42 IL6interleukin 6 (interferon, beta 2) (205207_at), score: 0.41 IL8interleukin 8 (211506_s_at), score: 0.36 JAM2junctional adhesion molecule 2 (219213_at), score: -0.78 JARID2jumonji, AT rich interactive domain 2 (203297_s_at), score: 0.34 JHDM1Djumonji C domain containing histone demethylase 1 homolog D (S. cerevisiae) (221778_at), score: 0.52 KCNMB4potassium large conductance calcium-activated channel, subfamily M, beta member 4 (219287_at), score: -0.4 KIAA1644KIAA1644 (52837_at), score: 0.54 LGALS3lectin, galactoside-binding, soluble, 3 (208949_s_at), score: 0.36 LGR4leucine-rich repeat-containing G protein-coupled receptor 4 (218326_s_at), score: -0.5 LHFPlipoma HMGIC fusion partner (218656_s_at), score: 0.56 LOXlysyl oxidase (213640_s_at), score: -0.43 LOXL2lysyl oxidase-like 2 (202997_s_at), score: -0.62 LY96lymphocyte antigen 96 (206584_at), score: 0.45 MAN1A1mannosidase, alpha, class 1A, member 1 (221760_at), score: 0.4 MAN1C1mannosidase, alpha, class 1C, member 1 (218918_at), score: 0.4 MDKmidkine (neurite growth-promoting factor 2) (209035_at), score: -0.39 MEG3maternally expressed 3 (non-protein coding) (210794_s_at), score: -0.39 MEOX1mesenchyme homeobox 1 (205619_s_at), score: -0.42 MESTmesoderm specific transcript homolog (mouse) (202016_at), score: -0.48 MSX1msh homeobox 1 (205932_s_at), score: 0.41 MYO1Dmyosin ID (212338_at), score: 0.58 NAMPTnicotinamide phosphoribosyltransferase (217738_at), score: 0.43 NAV2neuron navigator 2 (218330_s_at), score: 0.41 NFIL3nuclear factor, interleukin 3 regulated (203574_at), score: 0.45 NFKB1nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 (209239_at), score: 0.45 NPTX1neuronal pentraxin I (204684_at), score: 0.43 NR3C1nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) (201866_s_at), score: 0.35 NR4A3nuclear receptor subfamily 4, group A, member 3 (209959_at), score: 0.34 NT5DC25'-nucleotidase domain containing 2 (218051_s_at), score: -0.41 OCLMoculomedin (208274_at), score: -0.44 OLFML1olfactomedin-like 1 (217525_at), score: 0.45 OPCMLopioid binding protein/cell adhesion molecule-like (214111_at), score: -0.38 P4HA1prolyl 4-hydroxylase, alpha polypeptide I (207543_s_at), score: -0.57 PALMparalemmin (203859_s_at), score: -0.43 PAMpeptidylglycine alpha-amidating monooxygenase (214620_x_at), score: 0.41 PAPPA2pappalysin 2 (213332_at), score: -0.46 PCTK2PCTAIRE protein kinase 2 (221918_at), score: 0.35 PDGFRLplatelet-derived growth factor receptor-like (205226_at), score: 0.34 PFKFB36-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 (202464_s_at), score: 0.38 PFKLphosphofructokinase, liver (201102_s_at), score: -0.6 PHLDA1pleckstrin homology-like domain, family A, member 1 (217997_at), score: 0.4 PLA2G4Cphospholipase A2, group IVC (cytosolic, calcium-independent) (209785_s_at), score: 0.35 PNRC2proline-rich nuclear receptor coactivator 2 (217779_s_at), score: 0.44 POSTNperiostin, osteoblast specific factor (210809_s_at), score: -0.53 PPARDperoxisome proliferator-activated receptor delta (37152_at), score: 0.47 PPLperiplakin (203407_at), score: 0.66 PSAT1phosphoserine aminotransferase 1 (220892_s_at), score: -0.38 PSG4pregnancy specific beta-1-glycoprotein 4 (208191_x_at), score: 0.39 PSPHphosphoserine phosphatase (205048_s_at), score: 0.36 RAI14retinoic acid induced 14 (202052_s_at), score: -0.39 RAPGEF2Rap guanine nucleotide exchange factor (GEF) 2 (203097_s_at), score: 0.38 RASGRP1RAS guanyl releasing protein 1 (calcium and DAG-regulated) (205590_at), score: -0.39 RCAN2regulator of calcineurin 2 (203498_at), score: 0.56 RELBv-rel reticuloendotheliosis viral oncogene homolog B (205205_at), score: 0.41 RGS3regulator of G-protein signaling 3 (203823_at), score: 0.43 RRADRas-related associated with diabetes (204802_at), score: 0.56 SCRG1scrapie responsive protein 1 (205475_at), score: -0.57 SECTM1secreted and transmembrane 1 (213716_s_at), score: 0.48 SELENBP1selenium binding protein 1 (214433_s_at), score: 0.35 SFRP4secreted frizzled-related protein 4 (204051_s_at), score: -0.6 SFRS6splicing factor, arginine/serine-rich 6 (206108_s_at), score: 0.37 SH2B2SH2B adaptor protein 2 (205367_at), score: -0.39 SHBSrc homology 2 domain containing adaptor protein B (204657_s_at), score: 0.44 SLC35F2solute carrier family 35, member F2 (218826_at), score: 0.36 SLC39A8solute carrier family 39 (zinc transporter), member 8 (209267_s_at), score: 0.43 SMYD3SET and MYND domain containing 3 (218788_s_at), score: -0.43 SOD3superoxide dismutase 3, extracellular (205236_x_at), score: 0.65 SOX11SRY (sex determining region Y)-box 11 (204914_s_at), score: -0.62 STMN2stathmin-like 2 (203000_at), score: 0.41 SVEP1sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 (213247_at), score: 0.42 SVILsupervillin (202565_s_at), score: 0.34 SWAP70SWAP-70 protein (209307_at), score: 0.37 TGFBR3transforming growth factor, beta receptor III (204731_at), score: 0.34 THBDthrombomodulin (203887_s_at), score: 0.43 THBS3thrombospondin 3 (209561_at), score: -0.38 TIPARPTCDD-inducible poly(ADP-ribose) polymerase (212665_at), score: 0.42 TKTtransketolase (208699_x_at), score: 0.38 TNFAIP2tumor necrosis factor, alpha-induced protein 2 (202510_s_at), score: 0.51 TNFAIP8tumor necrosis factor, alpha-induced protein 8 (210260_s_at), score: 0.41 TNFRSF11Btumor necrosis factor receptor superfamily, member 11b (204933_s_at), score: 0.37 TNFRSF14tumor necrosis factor receptor superfamily, member 14 (herpesvirus entry mediator) (209354_at), score: 0.37 TNFRSF1Btumor necrosis factor receptor superfamily, member 1B (203508_at), score: 0.36 TNXAtenascin XA pseudogene (213451_x_at), score: 0.4 TNXBtenascin XB (216333_x_at), score: 0.49 TP53BP2tumor protein p53 binding protein, 2 (203120_at), score: 0.43 TPI1triosephosphate isomerase 1 (213011_s_at), score: -0.39 TRIB2tribbles homolog 2 (Drosophila) (202478_at), score: -0.38 TSC22D3TSC22 domain family, member 3 (208763_s_at), score: -0.4 UCHL1ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) (201387_s_at), score: -0.47 WDR41WD repeat domain 41 (218055_s_at), score: -0.41 WNT2wingless-type MMTV integration site family member 2 (205648_at), score: -0.53 WNT5Awingless-type MMTV integration site family, member 5A (213425_at), score: -0.38 WTAPWilms tumor 1 associated protein (210285_x_at), score: 0.36 YWHAHtyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide (201020_at), score: -0.39
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| t21c 08-03.CEL | 6 | 1 | DS-CC | hgu133a | Down | DS-CC 6 |
| t21b 08-03.CEL | 5 | 1 | DS-CC | hgu133a | Down | DS-CC 5 |
| t21d 08-03.CEL | 7 | 1 | DS-CC | hgu133a | Down | DS-CC 7 |
| ctrl b 08-03.CEL | 2 | 1 | DS-CC | hgu133a | none | DS-CC 2 |