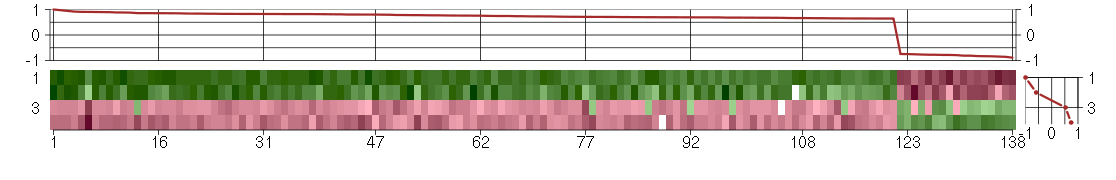

Under-expression is coded with green,
over-expression with red color.

biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
response to carbohydrate stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a carbohydrate stimulus.
response to hexose stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a hexose stimulus.
response to glucose stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a glucose stimulus.
response to organic substance
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an organic substance stimulus.
response to monosaccharide stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a monosaccharide stimulus.
response to chemical stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a chemical stimulus.
response to stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
all
This term is the most general term possible


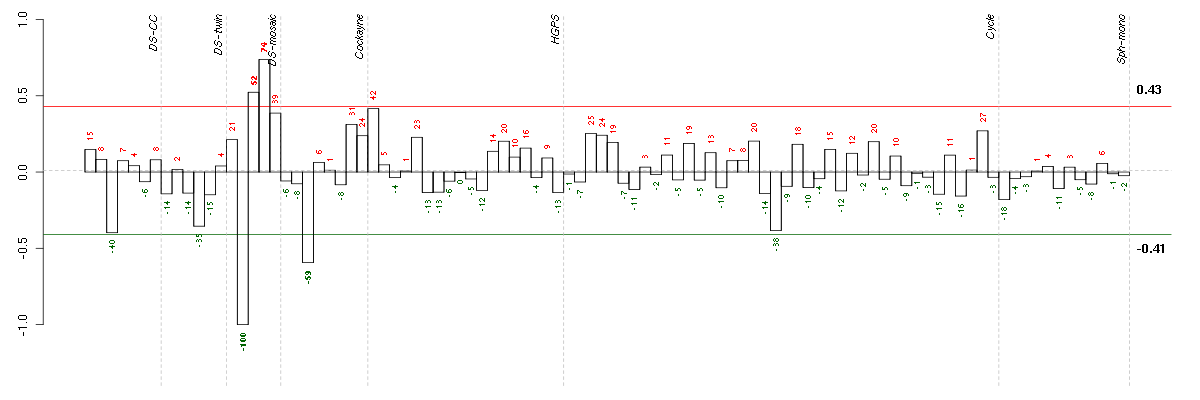
A4GALTalpha 1,4-galactosyltransferase (219488_at), score: 0.67 ADAM15ADAM metallopeptidase domain 15 (217007_s_at), score: 0.86 ADRBK1adrenergic, beta, receptor kinase 1 (201401_s_at), score: 0.84 AEBP1AE binding protein 1 (201792_at), score: 0.82 AP1S1adaptor-related protein complex 1, sigma 1 subunit (205195_at), score: 0.88 APOL6apolipoprotein L, 6 (219716_at), score: -0.86 ARFRP1ADP-ribosylation factor related protein 1 (215984_s_at), score: 0.74 ARHGDIARho GDP dissociation inhibitor (GDI) alpha (213606_s_at), score: 0.97 ARPC4actin related protein 2/3 complex, subunit 4, 20kDa (211672_s_at), score: 0.86 ATP13A2ATPase type 13A2 (218608_at), score: 0.65 BCL2L1BCL2-like 1 (215037_s_at), score: 0.83 BET1Lblocked early in transport 1 homolog (S. cerevisiae)-like (220470_at), score: 0.8 BRD2bromodomain containing 2 (214911_s_at), score: 0.7 BSGbasigin (Ok blood group) (208677_s_at), score: 0.71 C19orf22chromosome 19 open reading frame 22 (221764_at), score: 0.81 C19orf6chromosome 19 open reading frame 6 (213986_s_at), score: 0.86 C19orf61chromosome 19 open reading frame 61 (221335_x_at), score: 0.65 C21orf91chromosome 21 open reading frame 91 (220941_s_at), score: -0.78 CCND1cyclin D1 (208711_s_at), score: 0.82 CD44CD44 molecule (Indian blood group) (210916_s_at), score: 0.7 CDK10cyclin-dependent kinase 10 (210622_x_at), score: 0.71 CFIcomplement factor I (203854_at), score: -0.78 CHKAcholine kinase alpha (204233_s_at), score: 0.65 CHST5carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 5 (219182_at), score: -0.83 CLPTM1cleft lip and palate associated transmembrane protein 1 (211136_s_at), score: 0.82 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: 0.79 COL1A1collagen, type I, alpha 1 (217430_x_at), score: 0.9 COL6A1collagen, type VI, alpha 1 (212940_at), score: 0.68 COPAcoatomer protein complex, subunit alpha (214336_s_at), score: 0.77 DAPK3death-associated protein kinase 3 (203890_s_at), score: 1 DCTN5dynactin 5 (p25) (209231_s_at), score: 0.69 DDA1DET1 and DDB1 associated 1 (218260_at), score: 0.67 DENND3DENN/MADD domain containing 3 (212974_at), score: 0.65 DHX9DEAH (Asp-Glu-Ala-His) box polypeptide 9 (212105_s_at), score: 0.65 DSC3desmocollin 3 (206032_at), score: 0.91 DSG2desmoglein 2 (217901_at), score: 0.71 EGFepidermal growth factor (beta-urogastrone) (206254_at), score: -0.77 EIF2S1eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa (201143_s_at), score: 0.66 EIF4G1eukaryotic translation initiation factor 4 gamma, 1 (208624_s_at), score: 0.82 EIF5Aeukaryotic translation initiation factor 5A (201123_s_at), score: 0.83 ELK1ELK1, member of ETS oncogene family (210376_x_at), score: 0.77 EZRezrin (208621_s_at), score: 0.76 FGFR1fibroblast growth factor receptor 1 (210973_s_at), score: 0.83 FN1fibronectin 1 (214701_s_at), score: 0.73 FRMD4AFERM domain containing 4A (208476_s_at), score: 0.7 FSCN1fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus) (210933_s_at), score: 0.75 GLULglutamate-ammonia ligase (glutamine synthetase) (200648_s_at), score: 0.65 GPR137G protein-coupled receptor 137 (43934_at), score: 0.65 HIST2H2AA3histone cluster 2, H2aa3 (214290_s_at), score: 0.71 HMGA1high mobility group AT-hook 1 (210457_x_at), score: 0.83 HPS1Hermansky-Pudlak syndrome 1 (217354_s_at), score: 0.7 IL17RCinterleukin 17 receptor C (64440_at), score: 0.69 INHBEinhibin, beta E (210587_at), score: -0.85 IRF1interferon regulatory factor 1 (202531_at), score: -0.75 KCTD20potassium channel tetramerisation domain containing 20 (214849_at), score: 0.76 KDELR1KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1 (200922_at), score: 0.78 KIAA0100KIAA0100 (201729_s_at), score: 0.69 LAMP1lysosomal-associated membrane protein 1 (201551_s_at), score: 0.9 LOC100133105hypothetical protein LOC100133105 (214237_x_at), score: 0.73 LOXL2lysyl oxidase-like 2 (202997_s_at), score: 0.85 LSSlanosterol synthase (2,3-oxidosqualene-lanosterol cyclase) (211019_s_at), score: 0.77 LTBP3latent transforming growth factor beta binding protein 3 (219922_s_at), score: 0.68 MAFKv-maf musculoaponeurotic fibrosarcoma oncogene homolog K (avian) (206750_at), score: 0.75 MAP2K2mitogen-activated protein kinase kinase 2 (213490_s_at), score: 0.82 MAPKAPK2mitogen-activated protein kinase-activated protein kinase 2 (201461_s_at), score: 0.73 MAXMYC associated factor X (210734_x_at), score: 0.7 MAZMYC-associated zinc finger protein (purine-binding transcription factor) (207824_s_at), score: 0.75 MFN2mitofusin 2 (216205_s_at), score: 0.7 MGAT4Bmannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme B (220189_s_at), score: 0.72 MID2midline 2 (208384_s_at), score: -0.77 MLXMAX-like protein X (217909_s_at), score: 0.67 MMP14matrix metallopeptidase 14 (membrane-inserted) (202827_s_at), score: 0.95 MPDU1mannose-P-dolichol utilization defect 1 (209208_at), score: 0.78 NAB2NGFI-A binding protein 2 (EGR1 binding protein 2) (216017_s_at), score: 0.82 NAPAN-ethylmaleimide-sensitive factor attachment protein, alpha (206491_s_at), score: 0.69 NFATC4nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 (205897_at), score: 0.76 NFKB2nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) (207535_s_at), score: 0.7 NPTX1neuronal pentraxin I (204684_at), score: 0.66 NRBP1nuclear receptor binding protein 1 (217765_at), score: 0.71 NTMneurotrimin (222020_s_at), score: 0.8 NUCB1nucleobindin 1 (200646_s_at), score: 0.75 NUMA1nuclear mitotic apparatus protein 1 (214251_s_at), score: 0.85 OAZ2ornithine decarboxylase antizyme 2 (201364_s_at), score: 0.69 ORAI2ORAI calcium release-activated calcium modulator 2 (218812_s_at), score: 0.68 P4HBprolyl 4-hydroxylase, beta polypeptide (200656_s_at), score: 0.68 PACS1phosphofurin acidic cluster sorting protein 1 (220557_s_at), score: 0.65 PARP8poly (ADP-ribose) polymerase family, member 8 (219033_at), score: -0.81 PCDHGA11protocadherin gamma subfamily A, 11 (211876_x_at), score: 0.8 PCDHGA3protocadherin gamma subfamily A, 3 (216352_x_at), score: 0.85 PEX10peroxisomal biogenesis factor 10 (206351_s_at), score: 0.72 PFKLphosphofructokinase, liver (201102_s_at), score: 0.76 PIONpigeon homolog (Drosophila) (222150_s_at), score: -0.89 PIP5K1Aphosphatidylinositol-4-phosphate 5-kinase, type I, alpha (211205_x_at), score: 0.8 PLD3phospholipase D family, member 3 (201050_at), score: 0.73 PLEC1plectin 1, intermediate filament binding protein 500kDa (216971_s_at), score: 0.91 POLDIP3polymerase (DNA-directed), delta interacting protein 3 (215357_s_at), score: 0.88 POLR2Epolymerase (RNA) II (DNA directed) polypeptide E, 25kDa (213887_s_at), score: 0.68 PPP2R1Aprotein phosphatase 2 (formerly 2A), regulatory subunit A, alpha isoform (200695_at), score: 0.83 PPP5Cprotein phosphatase 5, catalytic subunit (201979_s_at), score: 0.78 PRPF6PRP6 pre-mRNA processing factor 6 homolog (S. cerevisiae) (208879_x_at), score: 0.84 PTBP1polypyrimidine tract binding protein 1 (212016_s_at), score: 0.92 PTMSparathymosin (218045_x_at), score: 0.84 PTRFpolymerase I and transcript release factor (208790_s_at), score: 0.9 PXNpaxillin (211823_s_at), score: 0.85 RAB11BRAB11B, member RAS oncogene family (34478_at), score: 0.69 RAB35RAB35, member RAS oncogene family (205461_at), score: 0.66 RAB3BRAB3B, member RAS oncogene family (205924_at), score: 0.66 RANGAP1Ran GTPase activating protein 1 (212125_at), score: 0.83 RASA3RAS p21 protein activator 3 (206220_s_at), score: 0.83 RASA4RAS p21 protein activator 4 (208534_s_at), score: 0.8 RELAv-rel reticuloendotheliosis viral oncogene homolog A (avian) (209878_s_at), score: 0.88 RNF126ring finger protein 126 (205748_s_at), score: 0.69 RUNDC3BRUN domain containing 3B (215321_at), score: -0.76 RXRBretinoid X receptor, beta (215099_s_at), score: 0.82 SAMD14sterile alpha motif domain containing 14 (213866_at), score: 0.68 SEMA5Asema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A (205405_at), score: 0.8 SH3GLB2SH3-domain GRB2-like endophilin B2 (218813_s_at), score: 0.83 SH3PXD2ASH3 and PX domains 2A (213252_at), score: 0.71 SHC1SHC (Src homology 2 domain containing) transforming protein 1 (201469_s_at), score: 0.82 SLC12A4solute carrier family 12 (potassium/chloride transporters), member 4 (209401_s_at), score: 0.82 SLC16A3solute carrier family 16, member 3 (monocarboxylic acid transporter 4) (202855_s_at), score: 0.76 SLC35E1solute carrier family 35, member E1 (222263_at), score: 0.74 SLC6A9solute carrier family 6 (neurotransmitter transporter, glycine), member 9 (207043_s_at), score: 0.78 SPPL2Bsignal peptide peptidase-like 2B (215833_s_at), score: 0.7 SRPRsignal recognition particle receptor (docking protein) (200917_s_at), score: 0.81 ST6GALNAC4ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 (221551_x_at), score: 0.68 STAT2signal transducer and activator of transcription 2, 113kDa (205170_at), score: 0.66 SULT1A2sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2 (207122_x_at), score: -0.81 TAF4TAF4 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 135kDa (213090_s_at), score: -0.83 TAPBPLTAP binding protein-like (218747_s_at), score: -0.75 TDRD3tudor domain containing 3 (208089_s_at), score: -0.77 TGFBR2transforming growth factor, beta receptor II (70/80kDa) (207334_s_at), score: 0.78 TLR3toll-like receptor 3 (206271_at), score: -0.84 TOP3Atopoisomerase (DNA) III alpha (214300_s_at), score: 0.65 UBE2Mubiquitin-conjugating enzyme E2M (UBC12 homolog, yeast) (203109_at), score: 0.71 UBTD1ubiquitin domain containing 1 (219172_at), score: 0.65 ZFPL1zinc finger protein-like 1 (209428_s_at), score: 0.82 ZRANB1zinc finger, RAN-binding domain containing 1 (201219_at), score: -0.79
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| 46B.CEL | 2 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 2 |
| E-GEOD-3407-raw-cel-1437949655.cel | 3 | 4 | Cockayne | hgu133a | none | CSB |
| 46C.CEL | 3 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 3 |
| 47B.CEL | 4 | 3 | DS-mosaic | hgu133plus2 | Down mosaic | DS-mosaic 4 |