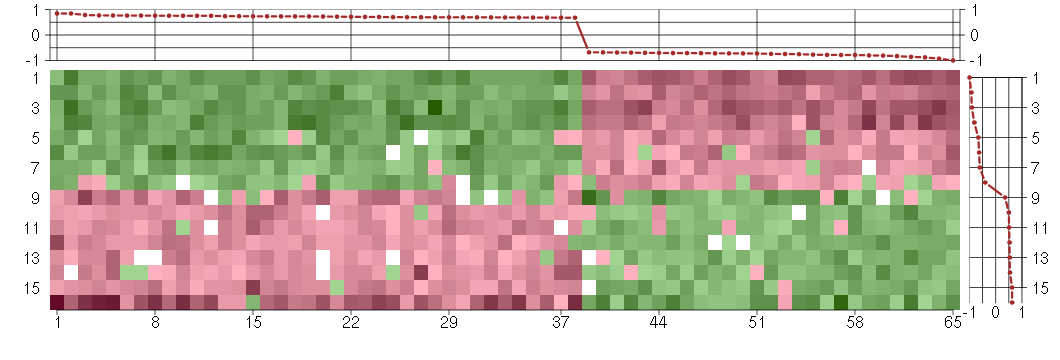

Under-expression is coded with green,
over-expression with red color.

immune system process
Any process involved in the development or functioning of the immune system, an organismal system for calibrated responses to potential internal or invasive threats.
immune system development
The process whose specific outcome is the progression of an organismal system whose objective is to provide calibrated responses by an organism to a potential internal or invasive threat, over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
leukocyte differentiation
The process whereby a relatively unspecialized hemopoietic precursor cell acquires the specialized features of a plasmacytoid dendritic cell or any cell of the myeloid leukocyte or lymphocyte lineages.
myeloid leukocyte differentiation
The process whereby a relatively unspecialized myeloid precursor cell acquires the specialized features of any cell of the myeloid leukocyte lineage.
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
hemopoiesis
The process whose specific outcome is the progression of the myeloid and lymphoid derived organ/tissue systems of the blood and other parts of the body over time, from formation to the mature structure. The site of hemopoiesis is variable during development, but occurs primarily in bone marrow or kidney in many adult vertebrates.
myeloid cell differentiation
The process whereby a relatively unspecialized myeloid precursor cell acquires the specialized features of any cell of the myeloid leukocyte, megakaryocyte, thrombocyte, or erythrocyte lineages.
cell differentiation
The process whereby relatively unspecialized cells, e.g. embryonic or regenerative cells, acquire specialized structural and/or functional features that characterize the cells, tissues, or organs of the mature organism or some other relatively stable phase of the organism's life history. Differentiation includes the processes involved in commitment of a cell to a specific fate.
osteoclast differentiation
The process whereby a relatively unspecialized monocyte acquires the specialized features of a osteoclast cell.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
developmental process
A biological process whose specific outcome is the progression of an integrated living unit: an anatomical structure (which may be a subcellular structure, cell, tissue, or organ), or organism over time from an initial condition to a later condition.
organ development
Development of a tissue or tissues that work together to perform a specific function or functions. Development pertains to the process whose specific outcome is the progression of a structure over time, from its formation to the mature structure. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
hemopoietic or lymphoid organ development
The process whose specific outcome is the progression of any organ involved in hemopoiesis or lymphoid cell activation over time, from its formation to the mature structure. Such development includes differentiation of resident cell types (stromal cells) and of migratory cell types dependent on the unique microenvironment afforded by the organ for their proper differentiation.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
anatomical structure development
The biological process whose specific outcome is the progression of an anatomical structure from an initial condition to its mature state. This process begins with the formation of the structure and ends with the mature structure, whatever form that may be including its natural destruction. An anatomical structure is any biological entity that occupies space and is distinguished from its surroundings. Anatomical structures can be macroscopic such as a carpel, or microscopic such as an acrosome.
cellular developmental process
A biological process whose specific outcome is the progression of a cell over time from an initial condition to a later condition.
all
This term is the most general term possible
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
cellular developmental process
A biological process whose specific outcome is the progression of a cell over time from an initial condition to a later condition.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
immune system development
The process whose specific outcome is the progression of an organismal system whose objective is to provide calibrated responses by an organism to a potential internal or invasive threat, over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
organ development
Development of a tissue or tissues that work together to perform a specific function or functions. Development pertains to the process whose specific outcome is the progression of a structure over time, from its formation to the mature structure. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
hemopoietic or lymphoid organ development
The process whose specific outcome is the progression of any organ involved in hemopoiesis or lymphoid cell activation over time, from its formation to the mature structure. Such development includes differentiation of resident cell types (stromal cells) and of migratory cell types dependent on the unique microenvironment afforded by the organ for their proper differentiation.
leukocyte differentiation
The process whereby a relatively unspecialized hemopoietic precursor cell acquires the specialized features of a plasmacytoid dendritic cell or any cell of the myeloid leukocyte or lymphocyte lineages.
myeloid cell differentiation
The process whereby a relatively unspecialized myeloid precursor cell acquires the specialized features of any cell of the myeloid leukocyte, megakaryocyte, thrombocyte, or erythrocyte lineages.
myeloid leukocyte differentiation
The process whereby a relatively unspecialized myeloid precursor cell acquires the specialized features of any cell of the myeloid leukocyte lineage.


molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
signal transducer activity
Mediates the transfer of a signal from the outside to the inside of a cell by means other than the introduction of the signal molecule itself into the cell.
receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity.
transmembrane receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity, and spanning to the membrane of either the cell or an organelle.
molecular transducer activity
The molecular function that accepts an input of one form and creates an output of a different form.
all
This term is the most general term possible
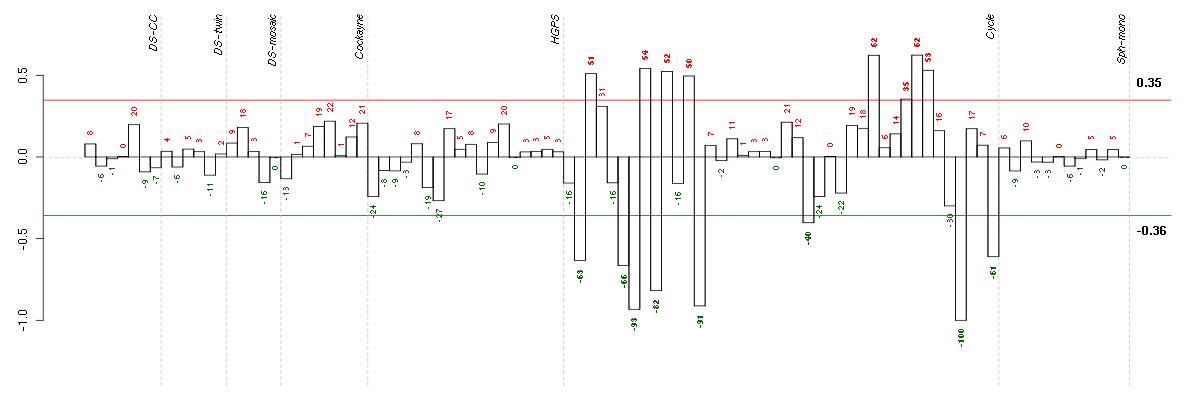
ACTR3BARP3 actin-related protein 3 homolog B (yeast) (218868_at), score: -0.72 ADORA2Badenosine A2b receptor (205891_at), score: 0.73 AIM1absent in melanoma 1 (212543_at), score: 0.73 ANXA10annexin A10 (210143_at), score: -0.68 BTBD2BTB (POZ) domain containing 2 (207722_s_at), score: 0.69 C4orf43chromosome 4 open reading frame 43 (218513_at), score: -0.7 CALB2calbindin 2 (205428_s_at), score: -0.78 CALCOCO1calcium binding and coiled-coil domain 1 (209002_s_at), score: 0.77 CDC42EP4CDC42 effector protein (Rho GTPase binding) 4 (218062_x_at), score: 0.68 CHUKconserved helix-loop-helix ubiquitous kinase (209666_s_at), score: -0.68 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: 0.84 DENND2ADENN/MADD domain containing 2A (53991_at), score: 0.69 DMPKdystrophia myotonica-protein kinase (37996_s_at), score: 0.75 DMWDdystrophia myotonica, WD repeat containing (33768_at), score: 0.69 DNAJB9DnaJ (Hsp40) homolog, subfamily B, member 9 (202843_at), score: -0.83 EPHA4EPH receptor A4 (206114_at), score: -0.71 FBXL14F-box and leucine-rich repeat protein 14 (213145_at), score: 0.69 GFOD1glucose-fructose oxidoreductase domain containing 1 (219821_s_at), score: -0.86 GK3Pglycerol kinase 3 pseudogene (215966_x_at), score: -0.8 GMCL1germ cell-less homolog 1 (Drosophila) (218458_at), score: -0.72 GTF2A1Lgeneral transcription factor IIA, 1-like (213413_at), score: 0.78 HSD17B6hydroxysteroid (17-beta) dehydrogenase 6 homolog (mouse) (37512_at), score: -0.78 HSPA12Aheat shock 70kDa protein 12A (214434_at), score: 0.72 JUPjunction plakoglobin (201015_s_at), score: 0.73 KAL1Kallmann syndrome 1 sequence (205206_at), score: -0.88 KIAA1305KIAA1305 (220911_s_at), score: 0.84 KLHL23kelch-like 23 (Drosophila) (213610_s_at), score: -0.75 LARP4La ribonucleoprotein domain family, member 4 (214155_s_at), score: -0.69 LOC100132540similar to LOC339047 protein (214870_x_at), score: 0.7 LOC339047hypothetical protein LOC339047 (221501_x_at), score: 0.72 LRP8low density lipoprotein receptor-related protein 8, apolipoprotein e receptor (205282_at), score: -0.81 MAN1C1mannosidase, alpha, class 1C, member 1 (218918_at), score: 0.75 MC4Rmelanocortin 4 receptor (221467_at), score: -0.7 MMP10matrix metallopeptidase 10 (stromelysin 2) (205680_at), score: -0.71 NEFMneurofilament, medium polypeptide (205113_at), score: -1 NPIPnuclear pore complex interacting protein (204538_x_at), score: 0.76 OAS22'-5'-oligoadenylate synthetase 2, 69/71kDa (204972_at), score: 0.77 OLFML2Bolfactomedin-like 2B (213125_at), score: 0.73 OPCMLopioid binding protein/cell adhesion molecule-like (214111_at), score: -0.75 OSTM1osteopetrosis associated transmembrane protein 1 (218196_at), score: -0.71 P2RY5purinergic receptor P2Y, G-protein coupled, 5 (218589_at), score: 0.76 PCDH9protocadherin 9 (219737_s_at), score: -0.93 PCYT2phosphate cytidylyltransferase 2, ethanolamine (209577_at), score: 0.73 PDE5Aphosphodiesterase 5A, cGMP-specific (206757_at), score: 0.69 PITX1paired-like homeodomain 1 (208502_s_at), score: 0.71 PPARGperoxisome proliferator-activated receptor gamma (208510_s_at), score: 0.69 RAB1ARAB1A, member RAS oncogene family (213440_at), score: -0.72 RASSF2Ras association (RalGDS/AF-6) domain family member 2 (203185_at), score: 0.68 REREarginine-glutamic acid dipeptide (RE) repeats (200940_s_at), score: 0.76 RNF220ring finger protein 220 (219988_s_at), score: 0.68 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: -0.69 SAV1salvador homolog 1 (Drosophila) (218276_s_at), score: 0.71 SCAMP4secretory carrier membrane protein 4 (213244_at), score: 0.74 SLC25A13solute carrier family 25, member 13 (citrin) (203775_at), score: -0.74 SLC4A7solute carrier family 4, sodium bicarbonate cotransporter, member 7 (209884_s_at), score: -0.77 SMAD3SMAD family member 3 (218284_at), score: 0.7 TENC1tensin like C1 domain containing phosphatase (tensin 2) (212494_at), score: 0.71 THSD7Athrombospondin, type I, domain containing 7A (214920_at), score: 0.68 TICAM2toll-like receptor adaptor molecule 2 (214658_at), score: -0.72 TLR3toll-like receptor 3 (206271_at), score: 0.7 TP53tumor protein p53 (201746_at), score: 0.75 TRIM21tripartite motif-containing 21 (204804_at), score: 0.75 ULBP2UL16 binding protein 2 (221291_at), score: -0.78 WDR6WD repeat domain 6 (217734_s_at), score: 0.74 ZDHHC7zinc finger, DHHC-type containing 7 (218606_at), score: 0.69
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486271.cel | 32 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486311.cel | 34 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485791.cel | 8 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486291.cel | 33 | 6 | Cycle | hgu133a2 | none | Cycle 1 |