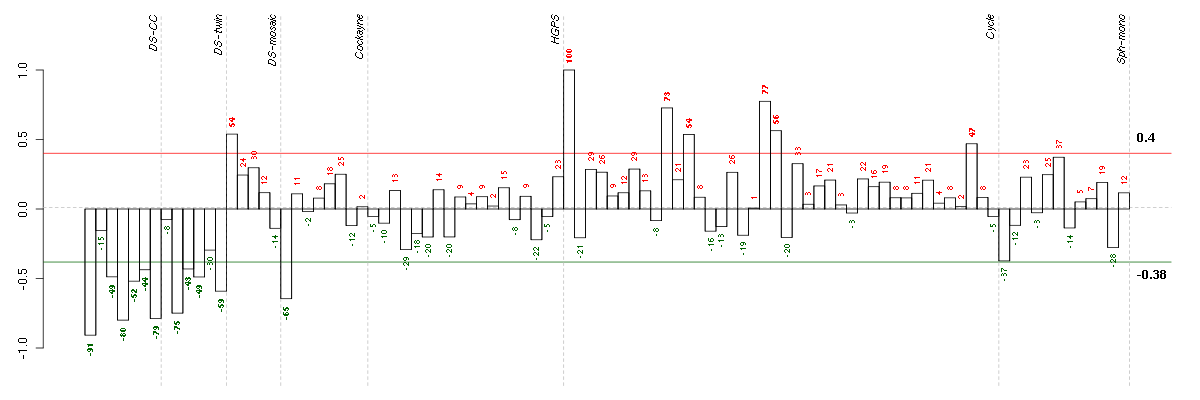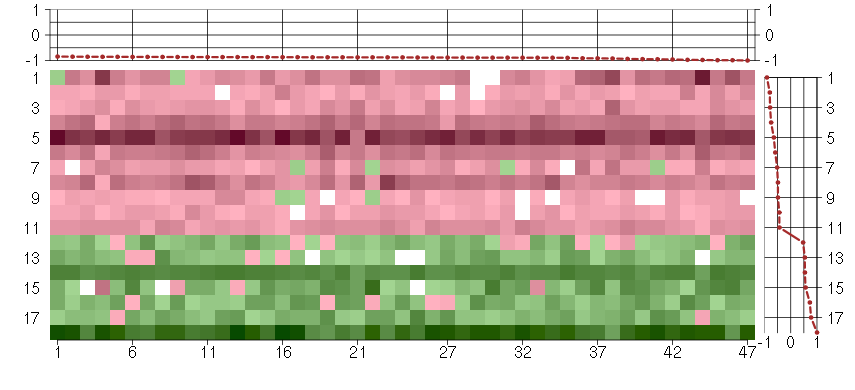

Under-expression is coded with green,
over-expression with red color.



molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
signal transducer activity
Mediates the transfer of a signal from the outside to the inside of a cell by means other than the introduction of the signal molecule itself into the cell.
receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity.
transmembrane receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity, and spanning to the membrane of either the cell or an organelle.
G-protein coupled receptor activity
A receptor that binds an extracellular ligand and transmits the signal to a heterotrimeric G-protein complex. These receptors are characteristically seven-transmembrane receptors and are made up of hetero- or homodimers.
molecular transducer activity
The molecular function that accepts an input of one form and creates an output of a different form.
all
This term is the most general term possible
ADAMTS9ADAM metallopeptidase with thrombospondin type 1 motif, 9 (220287_at), score: -0.87 AGMATagmatine ureohydrolase (agmatinase) (219792_at), score: -0.86 BEND5BEN domain containing 5 (219670_at), score: -0.88 BTNL3butyrophilin-like 3 (217207_s_at), score: -0.89 C1orf61chromosome 1 open reading frame 61 (205103_at), score: -0.89 C7orf28Achromosome 7 open reading frame 28A (201974_s_at), score: -0.99 C8orf17chromosome 8 open reading frame 17 (208266_at), score: -0.86 CASZ1castor zinc finger 1 (220015_at), score: -0.88 CEACAM6carcinoembryonic antigen-related cell adhesion molecule 6 (non-specific cross reacting antigen) (211657_at), score: -0.87 CLDN10claudin 10 (205328_at), score: -0.91 CLEC4MC-type lectin domain family 4, member M (207995_s_at), score: -0.88 CSN2casein beta (207951_at), score: -0.88 CTSEcathepsin E (205927_s_at), score: -0.89 CYP2A6cytochrome P450, family 2, subfamily A, polypeptide 6 (211295_x_at), score: -0.85 DAOD-amino-acid oxidase (206878_at), score: -0.89 DRD5dopamine receptor D5 (208486_at), score: -0.95 FAM38Bfamily with sequence similarity 38, member B (219602_s_at), score: -0.92 GNRHRgonadotropin-releasing hormone receptor (216341_s_at), score: -0.94 GPR12G protein-coupled receptor 12 (214558_at), score: -0.85 HAO2hydroxyacid oxidase 2 (long chain) (220801_s_at), score: -0.85 ITIH2inter-alpha (globulin) inhibitor H2 (204987_at), score: -0.88 KLF1Kruppel-like factor 1 (erythroid) (210504_at), score: -0.88 KRT2keratin 2 (207908_at), score: -0.87 LHX3LIM homeobox 3 (221670_s_at), score: -0.95 LOC389906similar to Serine/threonine-protein kinase PRKX (Protein kinase PKX1) (59433_at), score: -0.91 MBmyoglobin (204179_at), score: -0.89 MEP1Bmeprin A, beta (207251_at), score: -0.87 MFNGMFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase (204153_s_at), score: -0.87 MUC3Amucin 3A, cell surface associated (217117_x_at), score: -0.88 N4BP3Nedd4 binding protein 3 (214775_at), score: -0.88 NTNG1netrin G1 (206713_at), score: -0.86 OR7E24olfactory receptor, family 7, subfamily E, member 24 (215463_at), score: -0.96 PECAM1platelet/endothelial cell adhesion molecule (208982_at), score: -0.99 PEG3paternally expressed 3 (209242_at), score: -0.87 PLUNCpalate, lung and nasal epithelium associated (220542_s_at), score: -0.86 PRSS7protease, serine, 7 (enterokinase) (217269_s_at), score: -0.86 PYHIN1pyrin and HIN domain family, member 1 (216748_at), score: -0.86 RETret proto-oncogene (205879_x_at), score: -0.9 RP11-35N6.1plasticity related gene 3 (219732_at), score: -0.87 RPGRIP1retinitis pigmentosa GTPase regulator interacting protein 1 (206608_s_at), score: -1 RRHretinal pigment epithelium-derived rhodopsin homolog (208314_at), score: -0.87 S100A14S100 calcium binding protein A14 (218677_at), score: -0.89 SLC10A1solute carrier family 10 (sodium/bile acid cotransporter family), member 1 (207185_at), score: -0.89 TAS2R9taste receptor, type 2, member 9 (221461_at), score: -0.88 TBX21T-box 21 (220684_at), score: -0.88 TRA@T cell receptor alpha locus (216540_at), score: -0.97 VGFVGF nerve growth factor inducible (205586_x_at), score: -0.98
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| ctrl a 08-03.CEL | 1 | 1 | DS-CC | hgu133a | none | DS-CC 1 |
| t21a 08-03.CEL | 4 | 1 | DS-CC | hgu133a | Down | DS-CC 4 |
| t21d 08-03.CEL | 7 | 1 | DS-CC | hgu133a | Down | DS-CC 7 |
| 2Twin.CEL | 2 | 2 | DS-twin | hgu133plus2 | none | DS-twin 2 |
| E-GEOD-3407-raw-cel-1437949557.cel | 1 | 4 | Cockayne | hgu133a | CS | eGFP |
| 6Twin.CEL | 6 | 2 | DS-twin | hgu133plus2 | none | DS-twin 6 |
| t21b 08-03.CEL | 5 | 1 | DS-CC | hgu133a | Down | DS-CC 5 |
| 4Twin.CEL | 4 | 2 | DS-twin | hgu133plus2 | none | DS-twin 4 |
| ctrl c 08-03.CEL | 3 | 1 | DS-CC | hgu133a | none | DS-CC 3 |
| t21c 08-03.CEL | 6 | 1 | DS-CC | hgu133a | Down | DS-CC 6 |
| 3Twin.CEL | 3 | 2 | DS-twin | hgu133plus2 | Down | DS-twin 3 |
| E-TABM-263-raw-cel-1515486391.cel | 38 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 46A.CEL | 1 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 1 |
| E-TABM-263-raw-cel-1515486031.cel | 20 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486011.cel | 19 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485651.cel | 1 | 6 | Cycle | hgu133a2 | none | Cycle 1 |