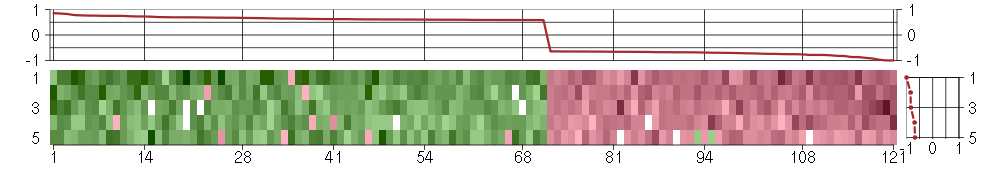

Under-expression is coded with green,
over-expression with red color.

MAPKKK cascade
Cascade of at least three protein kinase activities culminating in the phosphorylation and activation of a MAP kinase. MAPKKK cascades lie downstream of numerous signaling pathways.
signal transduction
The cascade of processes by which a signal interacts with a receptor, causing a change in the level or activity of a second messenger or other downstream target, and ultimately effecting a change in the functioning of the cell.
cell communication
Any process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
intracellular signaling cascade
A series of reactions within the cell that occur as a result of a single trigger reaction or compound.
protein kinase cascade
A series of reactions, mediated by protein kinases, which occurs as a result of a single trigger reaction or compound.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
positive regulation of kinase activity
Any process that activates or increases the frequency, rate or extent of kinase activity, the catalysis of the transfer of a phosphate group, usually from ATP, to a substrate molecule.
positive regulation of catalytic activity
Any process that activates or increases the activity of an enzyme.
regulation of MAP kinase activity
Any process that modulates the frequency, rate or extent of MAP kinase activity.
positive regulation of MAP kinase activity
Any process that activates or increases the frequency, rate or extent of MAP kinase activity.
regulation of JUN kinase activity
Any process that modulates the frequency, rate or extent of JUN kinase activity.
positive regulation of JUN kinase activity
Any process that activates or increases the frequency, rate or extent of JUN kinase activity.
regulation of kinase activity
Any process that modulates the frequency, rate or extent of kinase activity, the catalysis of the transfer of a phosphate group, usually from ATP, to a substrate molecule.
regulation of protein kinase activity
Any process that modulates the frequency, rate or extent of protein kinase activity.
positive regulation of protein kinase activity
Any process that activates or increases the frequency, rate or extent of protein kinase activity.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of catalytic activity
Any process that modulates the activity of an enzyme.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of transferase activity
Any process that modulates the frequency, rate or extent of transferase activity, the catalysis of the transfer of a group, e.g. a methyl group, glycosyl group, acyl group, phosphorus-containing, or other groups, from one compound (generally regarded as the donor) to another compound (generally regarded as the acceptor). Transferase is the systematic name for any enzyme of EC class 2.
positive regulation of transferase activity
Any process that activates or increases the frequency, rate or extent of transferase activity, the catalysis of the transfer of a group, e.g. a methyl group, glycosyl group, acyl group, phosphorus-containing, or other groups, from a donor compound to an acceptor.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
regulation of molecular function
Any process that modulates the frequency, rate or extent of molecular functions. Molecular functions are elemental biological activities occurring at the molecular level, such as catalysis or binding.
all
This term is the most general term possible
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
signal transduction
The cascade of processes by which a signal interacts with a receptor, causing a change in the level or activity of a second messenger or other downstream target, and ultimately effecting a change in the functioning of the cell.
positive regulation of transferase activity
Any process that activates or increases the frequency, rate or extent of transferase activity, the catalysis of the transfer of a group, e.g. a methyl group, glycosyl group, acyl group, phosphorus-containing, or other groups, from a donor compound to an acceptor.
positive regulation of kinase activity
Any process that activates or increases the frequency, rate or extent of kinase activity, the catalysis of the transfer of a phosphate group, usually from ATP, to a substrate molecule.
positive regulation of protein kinase activity
Any process that activates or increases the frequency, rate or extent of protein kinase activity.
positive regulation of MAP kinase activity
Any process that activates or increases the frequency, rate or extent of MAP kinase activity.
positive regulation of JUN kinase activity
Any process that activates or increases the frequency, rate or extent of JUN kinase activity.


molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
signal transducer activity
Mediates the transfer of a signal from the outside to the inside of a cell by means other than the introduction of the signal molecule itself into the cell.
receptor signaling protein activity
NA
molecular transducer activity
The molecular function that accepts an input of one form and creates an output of a different form.
all
This term is the most general term possible
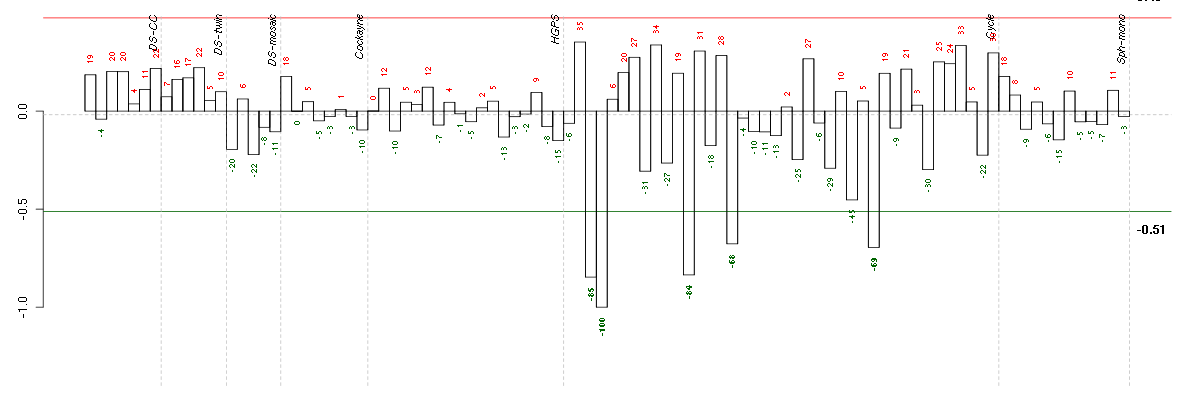
ACVR2Aactivin A receptor, type IIA (205327_s_at), score: 0.6 AGTangiotensinogen (serpin peptidase inhibitor, clade A, member 8) (202834_at), score: 0.61 ALDH1L1aldehyde dehydrogenase 1 family, member L1 (215798_at), score: 0.76 ALDH3A1aldehyde dehydrogenase 3 family, memberA1 (205623_at), score: 0.63 ARHGDIBRho GDP dissociation inhibitor (GDI) beta (201288_at), score: -0.64 ARL8BADP-ribosylation factor-like 8B (217852_s_at), score: 0.62 BAT2HLA-B associated transcript 2 (212081_x_at), score: -0.68 BAT2LHLA-B associated transcript 2-like (212068_s_at), score: -0.65 C14orf94chromosome 14 open reading frame 94 (218383_at), score: -0.66 C15orf5chromosome 15 open reading frame 5 (208109_s_at), score: 0.64 C19orf61chromosome 19 open reading frame 61 (221335_x_at), score: -0.69 C2orf37chromosome 2 open reading frame 37 (220172_at), score: -0.71 C7orf58chromosome 7 open reading frame 58 (220032_at), score: -0.72 CALB2calbindin 2 (205428_s_at), score: 0.76 CCDC94coiled-coil domain containing 94 (204335_at), score: 0.6 CD79BCD79b molecule, immunoglobulin-associated beta (205297_s_at), score: 0.61 CDC42EP4CDC42 effector protein (Rho GTPase binding) 4 (218062_x_at), score: -0.64 CDKN2Dcyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) (210240_s_at), score: -0.65 CMAHcytidine monophosphate-N-acetylneuraminic acid hydroxylase (CMP-N-acetylneuraminate monooxygenase) pseudogene (205518_s_at), score: -0.66 CNNM1cyclin M1 (220166_at), score: 0.58 CPNE3copine III (202118_s_at), score: -0.65 CXorf21chromosome X open reading frame 21 (220252_x_at), score: 0.59 CYP3A4cytochrome P450, family 3, subfamily A, polypeptide 4 (205998_x_at), score: 0.86 DENND3DENN/MADD domain containing 3 (212974_at), score: -0.68 DMBT1deleted in malignant brain tumors 1 (208250_s_at), score: 0.75 DNAJC2DnaJ (Hsp40) homolog, subfamily C, member 2 (213097_s_at), score: 0.6 DNASE1L3deoxyribonuclease I-like 3 (205554_s_at), score: 0.59 DSPPdentin sialophosphoprotein (221681_s_at), score: 0.59 EMCNendomucin (219436_s_at), score: -1 EPAGearly lymphoid activation protein (217050_at), score: 0.61 ERCC6excision repair cross-complementing rodent repair deficiency, complementation group 6 (207347_at), score: -0.67 F11coagulation factor XI (206610_s_at), score: 0.61 FAT1FAT tumor suppressor homolog 1 (Drosophila) (201579_at), score: 0.59 FBP1fructose-1,6-bisphosphatase 1 (209696_at), score: 0.68 FCER1GFc fragment of IgE, high affinity I, receptor for; gamma polypeptide (204232_at), score: 0.59 FGL1fibrinogen-like 1 (205305_at), score: 0.62 FLJ21075hypothetical protein FLJ21075 (221172_at), score: 0.77 FLRT3fibronectin leucine rich transmembrane protein 3 (219250_s_at), score: -0.87 GABARAPL1GABA(A) receptor-associated protein like 1 (208868_s_at), score: 0.68 GABARAPL3GABA(A) receptors associated protein like 3 (pseudogene) (211458_s_at), score: 0.75 GADD45Bgrowth arrest and DNA-damage-inducible, beta (207574_s_at), score: 0.59 GBA3glucosidase, beta, acid 3 (cytosolic) (219954_s_at), score: 0.73 GDF9growth differentiation factor 9 (221314_at), score: 0.6 GFOD1glucose-fructose oxidoreductase domain containing 1 (219821_s_at), score: 0.64 GMPRguanosine monophosphate reductase (204187_at), score: -0.66 GNL3Lguanine nucleotide binding protein-like 3 (nucleolar)-like (205010_at), score: 0.67 GOLGA8Ggolgi autoantigen, golgin subfamily a, 8G (222149_x_at), score: 0.66 GPR144G protein-coupled receptor 144 (216289_at), score: 0.72 GPR32G protein-coupled receptor 32 (221469_at), score: 0.69 GPR56G protein-coupled receptor 56 (212070_at), score: -0.75 HAB1B1 for mucin (215778_x_at), score: 0.69 HAND1heart and neural crest derivatives expressed 1 (220138_at), score: 0.69 HIST1H1Chistone cluster 1, H1c (209398_at), score: -0.68 HMGA1high mobility group AT-hook 1 (210457_x_at), score: -0.76 IDI2isopentenyl-diphosphate delta isomerase 2 (217631_at), score: 0.59 IL9Rinterleukin 9 receptor (217212_s_at), score: 0.69 INTS1integrator complex subunit 1 (212212_s_at), score: -0.66 JAK2Janus kinase 2 (a protein tyrosine kinase) (205842_s_at), score: -0.68 JUPjunction plakoglobin (201015_s_at), score: -0.74 KCTD15potassium channel tetramerisation domain containing 15 (218553_s_at), score: 0.59 KIF26Bkinesin family member 26B (220002_at), score: 0.6 KLF7Kruppel-like factor 7 (ubiquitous) (204334_at), score: 0.6 LEPREL2leprecan-like 2 (204854_at), score: -0.74 LMO2LIM domain only 2 (rhombotin-like 1) (204249_s_at), score: -0.66 LOC391132similar to hCG2041276 (216177_at), score: 0.62 LOC81691exonuclease NEF-sp (208107_s_at), score: -0.7 LRDDleucine-rich repeats and death domain containing (221640_s_at), score: -0.8 LRP2low density lipoprotein-related protein 2 (205710_at), score: -0.66 MAGEB2melanoma antigen family B, 2 (206218_at), score: 0.7 MAGOH2mago-nashi homolog 2, proliferation-associated (Drosophila) (217692_at), score: 0.61 MAP3K2mitogen-activated protein kinase kinase kinase 2 (221695_s_at), score: -0.65 MAP3K6mitogen-activated protein kinase kinase kinase 6 (219278_at), score: -0.78 MAPK10mitogen-activated protein kinase 10 (204813_at), score: 0.64 MEOX1mesenchyme homeobox 1 (205619_s_at), score: 0.73 MOBPmyelin-associated oligodendrocyte basic protein (210193_at), score: 0.59 MOGmyelin oligodendrocyte glycoprotein (214650_x_at), score: 0.75 MRPS6mitochondrial ribosomal protein S6 (213167_s_at), score: -0.73 MTMR11myotubularin related protein 11 (205076_s_at), score: -0.82 MYL10myosin, light chain 10, regulatory (221659_s_at), score: 0.64 NCOA2nuclear receptor coactivator 2 (205732_s_at), score: -0.67 NCRNA00093non-protein coding RNA 93 (210723_x_at), score: 0.6 NDPNorrie disease (pseudoglioma) (206022_at), score: -0.78 NF1neurofibromin 1 (211094_s_at), score: -0.69 OAS32'-5'-oligoadenylate synthetase 3, 100kDa (218400_at), score: -0.65 OLFML2Bolfactomedin-like 2B (213125_at), score: -1 OLR1oxidized low density lipoprotein (lectin-like) receptor 1 (210004_at), score: 0.68 PAK1p21 protein (Cdc42/Rac)-activated kinase 1 (209615_s_at), score: -0.9 PCDH24protocadherin 24 (220186_s_at), score: 0.62 PDE5Aphosphodiesterase 5A, cGMP-specific (206757_at), score: -0.82 PHF7PHD finger protein 7 (215622_x_at), score: 0.66 PNMAL1PNMA-like 1 (218824_at), score: -0.7 POU4F1POU class 4 homeobox 1 (206940_s_at), score: 0.59 PPP1R13Bprotein phosphatase 1, regulatory (inhibitor) subunit 13B (216347_s_at), score: 0.75 PRRX1paired related homeobox 1 (205991_s_at), score: -0.68 PTPN6protein tyrosine phosphatase, non-receptor type 6 (206687_s_at), score: 0.78 PXNpaxillin (211823_s_at), score: -0.78 RASA4RAS p21 protein activator 4 (208534_s_at), score: -0.65 RASGRP1RAS guanyl releasing protein 1 (calcium and DAG-regulated) (205590_at), score: 0.62 RASGRP3RAS guanyl releasing protein 3 (calcium and DAG-regulated) (205801_s_at), score: -0.97 REG3Aregenerating islet-derived 3 alpha (205815_at), score: 0.68 SAA4serum amyloid A4, constitutive (207096_at), score: 0.61 SDPRserum deprivation response (phosphatidylserine binding protein) (218711_s_at), score: -0.92 SLC1A7solute carrier family 1 (glutamate transporter), member 7 (210923_at), score: 0.69 SLC26A10solute carrier family 26, member 10 (214951_at), score: 0.69 SLC29A1solute carrier family 29 (nucleoside transporters), member 1 (201801_s_at), score: -0.7 SLC2A3solute carrier family 2 (facilitated glucose transporter), member 3 (202499_s_at), score: 0.65 SNTG1syntrophin, gamma 1 (220405_at), score: -0.75 SORBS1sorbin and SH3 domain containing 1 (218087_s_at), score: 0.61 SPATA1spermatogenesis associated 1 (221057_at), score: 0.65 ST6GAL1ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 (201998_at), score: 0.6 SULT1B1sulfotransferase family, cytosolic, 1B, member 1 (207601_at), score: 0.82 TAL1T-cell acute lymphocytic leukemia 1 (206283_s_at), score: 0.63 TGFBR2transforming growth factor, beta receptor II (70/80kDa) (207334_s_at), score: -0.87 TGM4transglutaminase 4 (prostate) (217566_s_at), score: 0.7 TOM1L2target of myb1-like 2 (chicken) (214840_at), score: -0.75 TRAPPC6Atrafficking protein particle complex 6A (204985_s_at), score: 0.61 TULP2tubby like protein 2 (206733_at), score: 0.73 VEGFBvascular endothelial growth factor B (203683_s_at), score: -0.72 ZNF185zinc finger protein 185 (LIM domain) (203585_at), score: -0.65 ZNF652zinc finger protein 652 (205594_at), score: 0.84
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485951.cel | 16 | 6 | Cycle | hgu133a2 | none | Cycle 1 |