
Under-expression is coded with green,
over-expression with red color.



molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
transporter activity
Enables the directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells.
transmembrane transporter activity
Catalysis of the transfer of a substance from one side of a membrane to the other.
organic acid:sodium symporter activity
Catalysis of the transfer of a solute or solutes from one side of a membrane to the other according to the reaction: organic acid(out) + Na+(out) = organic acid(in) + Na+(in).
organic acid transmembrane transporter activity
Catalysis of the transfer of organic acids, any acidic compound containing carbon in covalent linkage, from one side of the membrane to the other.
secondary active transmembrane transporter activity
Catalysis of the transfer of a solute from one side of a membrane to the other, up its concentration gradient. The transporter binds the solute and undergoes a series of conformational changes. Transport works equally well in either direction and is driven by a chemiosmotic source of energy. Chemiosmotic sources of energy include uniport, symport or antiport.
monocarboxylic acid transmembrane transporter activity
Catalysis of the transfer of monocarboxylic acids from one side of the membrane to the other. A monocarboxylic acid is an organic acid with one COOH group.
cation transmembrane transporter activity
Catalysis of the transfer of cation from one side of the membrane to the other.
bile acid:sodium symporter activity
Catalysis of the transfer of a solute or solutes from one side of a membrane to the other according to the reaction: bile acid(out) + Na+(out) = bile acid(in) + Na+(in).
ion transmembrane transporter activity
Catalysis of the transfer of an ion from one side of a membrane to the other.
bile acid transmembrane transporter activity
Catalysis of the transfer of bile acid from one side of the membrane to the other. Bile acids are any of a group of steroid carboxylic acids occurring in bile, where they are present as the sodium salts of their amides with glycine or taurine.
symporter activity
Enables the active transport of a solute across a membrane by a mechanism whereby two or more species are transported together in the same direction in a tightly coupled process not directly linked to a form of energy other than chemiosmotic energy.
solute:cation symporter activity
Catalysis of the transfer of a solute or solutes from one side of a membrane to the other according to the reaction: solute(out) + cation(out) = solute(in) + cation(in).
solute:sodium symporter activity
Catalysis of the transfer of a solute or solutes from one side of a membrane to the other according to the reaction: solute(out) + Na+(out) = solute(in) + Na+(in).
active transmembrane transporter activity
Catalysis of the transfer of a specific substance or related group of substances from one side of a membrane to the other, up the solute's concentration gradient. The transporter binds the solute and undergoes a series of conformational changes. Transport works equally well in either direction.
substrate-specific transmembrane transporter activity
Catalysis of the transfer of a specific substance or group of related substances from one side of a membrane to the other.
substrate-specific transporter activity
Enables the directed movement of a specific substance or group of related substances (such as macromolecules, small molecules, ions) into, out of, within or between cells.
carboxylic acid transmembrane transporter activity
Catalysis of the transfer of carboxylic acids from one side of the membrane to the other. Carboxylic acids are organic acids containing one or more carboxyl (COOH) groups or anions (COO-).
all
This term is the most general term possible
substrate-specific transmembrane transporter activity
Catalysis of the transfer of a specific substance or group of related substances from one side of a membrane to the other.
solute:cation symporter activity
Catalysis of the transfer of a solute or solutes from one side of a membrane to the other according to the reaction: solute(out) + cation(out) = solute(in) + cation(in).
organic acid:sodium symporter activity
Catalysis of the transfer of a solute or solutes from one side of a membrane to the other according to the reaction: organic acid(out) + Na+(out) = organic acid(in) + Na+(in).
bile acid:sodium symporter activity
Catalysis of the transfer of a solute or solutes from one side of a membrane to the other according to the reaction: bile acid(out) + Na+(out) = bile acid(in) + Na+(in).
A1CFAPOBEC1 complementation factor (220951_s_at), score: -0.51 ABOABO blood group (transferase A, alpha 1-3-N-acetylgalactosaminyltransferase; transferase B, alpha 1-3-galactosyltransferase) (216716_at), score: -0.55 ADH1Balcohol dehydrogenase 1B (class I), beta polypeptide (209612_s_at), score: -0.65 ADRA2Aadrenergic, alpha-2A-, receptor (209869_at), score: -0.85 APODapolipoprotein D (201525_at), score: -0.68 ASAP3ArfGAP with SH3 domain, ankyrin repeat and PH domain 3 (219103_at), score: -0.52 BTNL3butyrophilin-like 3 (217207_s_at), score: -0.54 C7orf28Achromosome 7 open reading frame 28A (201974_s_at), score: -0.57 CATSPER2cation channel, sperm associated 2 (217588_at), score: -0.5 CCDC9coiled-coil domain containing 9 (206257_at), score: -0.54 CEACAM6carcinoembryonic antigen-related cell adhesion molecule 6 (non-specific cross reacting antigen) (211657_at), score: -0.5 CLDN10claudin 10 (205328_at), score: -0.55 CTSDcathepsin D (200766_at), score: -0.5 CTSFcathepsin F (203657_s_at), score: -0.58 CYP3A4cytochrome P450, family 3, subfamily A, polypeptide 4 (205998_x_at), score: -0.55 DKK2dickkopf homolog 2 (Xenopus laevis) (219908_at), score: -1 DPTdermatopontin (213068_at), score: -0.67 DRD5dopamine receptor D5 (208486_at), score: -0.57 ENPP4ectonucleotide pyrophosphatase/phosphodiesterase 4 (putative function) (204160_s_at), score: -0.54 FAM38Bfamily with sequence similarity 38, member B (219602_s_at), score: -0.52 FBLN2fibulin 2 (203886_s_at), score: -0.51 FOSBFBJ murine osteosarcoma viral oncogene homolog B (202768_at), score: -0.7 GAAglucosidase, alpha; acid (202812_at), score: -0.56 GPATCH4G patch domain containing 4 (220596_at), score: -0.49 GPR12G protein-coupled receptor 12 (214558_at), score: -0.58 GRIA1glutamate receptor, ionotropic, AMPA 1 (209793_at), score: -0.77 HIVEP2human immunodeficiency virus type I enhancer binding protein 2 (212642_s_at), score: -0.57 JMJD3jumonji domain containing 3, histone lysine demethylase (213146_at), score: -0.65 KCNJ16potassium inwardly-rectifying channel, subfamily J, member 16 (219564_at), score: -0.5 KIR3DX1killer cell immunoglobulin-like receptor, three domains, X1 (216428_x_at), score: -0.52 KLF1Kruppel-like factor 1 (erythroid) (210504_at), score: -0.5 LHX3LIM homeobox 3 (221670_s_at), score: -0.54 LOC100188945cell division cycle associated 4 pseudogene (215109_at), score: -0.58 LOC389906similar to Serine/threonine-protein kinase PRKX (Protein kinase PKX1) (59433_at), score: -0.56 LOC80054hypothetical LOC80054 (220465_at), score: -0.51 LOH3CR2Aloss of heterozygosity, 3, chromosomal region 2, gene A (220244_at), score: -0.83 LONRF3LON peptidase N-terminal domain and ring finger 3 (220009_at), score: -0.53 LRRN2leucine rich repeat neuronal 2 (216164_at), score: -0.49 MAPK8IP2mitogen-activated protein kinase 8 interacting protein 2 (205050_s_at), score: -0.57 MFNGMFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase (204153_s_at), score: -0.51 MMP9matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) (203936_s_at), score: -0.5 MMRN2multimerin 2 (219091_s_at), score: -0.75 MOBPmyelin-associated oligodendrocyte basic protein (210193_at), score: -0.67 MYL4myosin, light chain 4, alkali; atrial, embryonic (210088_x_at), score: -0.52 MYO1Amyosin IA (211916_s_at), score: -0.77 N4BP3Nedd4 binding protein 3 (214775_at), score: -0.5 NRN1neuritin 1 (218625_at), score: -0.65 OR1E2olfactory receptor, family 1, subfamily E, member 2 (208587_s_at), score: -0.63 OR7E24olfactory receptor, family 7, subfamily E, member 24 (215463_at), score: -0.6 PECAM1platelet/endothelial cell adhesion molecule (208982_at), score: -0.59 PGK2phosphoglycerate kinase 2 (217009_at), score: -0.55 PIPOXpipecolic acid oxidase (221605_s_at), score: -0.53 PNPLA2patatin-like phospholipase domain containing 2 (212705_x_at), score: -0.75 PRSS7protease, serine, 7 (enterokinase) (217269_s_at), score: -0.83 PTPREprotein tyrosine phosphatase, receptor type, E (221840_at), score: -0.76 PYHIN1pyrin and HIN domain family, member 1 (216748_at), score: -0.54 RASL11BRAS-like, family 11, member B (219142_at), score: -0.64 RP11-35N6.1plasticity related gene 3 (219732_at), score: -0.57 RP3-377H14.5hypothetical LOC285830 (222279_at), score: -0.57 RPGRIP1retinitis pigmentosa GTPase regulator interacting protein 1 (206608_s_at), score: -0.58 RRADRas-related associated with diabetes (204802_at), score: -0.75 RUNX3runt-related transcription factor 3 (204198_s_at), score: -0.56 S100A14S100 calcium binding protein A14 (218677_at), score: -0.5 SCN10Asodium channel, voltage-gated, type X, alpha subunit (208578_at), score: -0.53 SLC10A1solute carrier family 10 (sodium/bile acid cotransporter family), member 1 (207185_at), score: -0.55 SLC10A2solute carrier family 10 (sodium/bile acid cotransporter family), member 2 (207095_at), score: -0.49 SOCS3suppressor of cytokine signaling 3 (206359_at), score: -0.53 SRCAPSnf2-related CREBBP activator protein (212275_s_at), score: -0.51 ST6GAL1ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 (201998_at), score: -0.52 TACR1tachykinin receptor 1 (208048_at), score: -0.68 TBX21T-box 21 (220684_at), score: -0.54 TBX6T-box 6 (207684_at), score: -0.57 TRA@T cell receptor alpha locus (216540_at), score: -0.66 VGFVGF nerve growth factor inducible (205586_x_at), score: -0.86
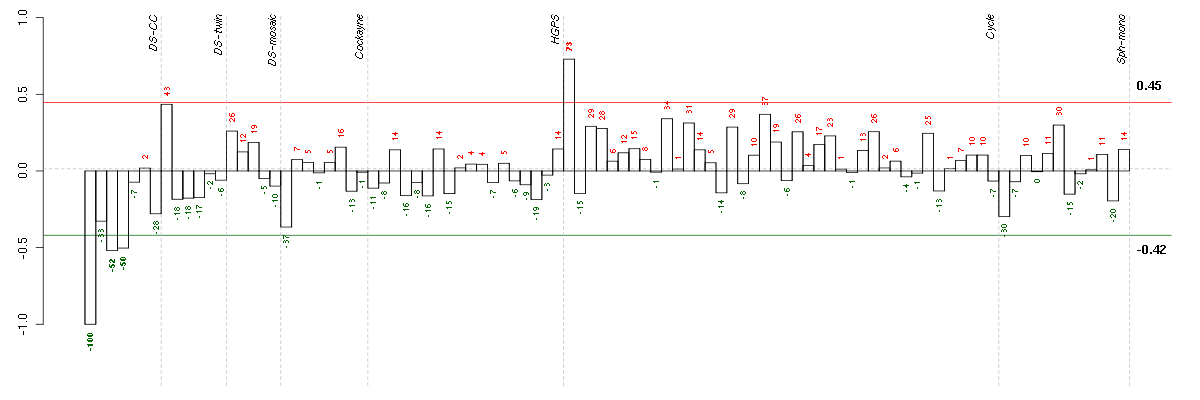
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| ctrl a 08-03.CEL | 1 | 1 | DS-CC | hgu133a | none | DS-CC 1 |
| ctrl c 08-03.CEL | 3 | 1 | DS-CC | hgu133a | none | DS-CC 3 |
| t21a 08-03.CEL | 4 | 1 | DS-CC | hgu133a | Down | DS-CC 4 |
| E-TABM-263-raw-cel-1515485651.cel | 1 | 6 | Cycle | hgu133a2 | none | Cycle 1 |