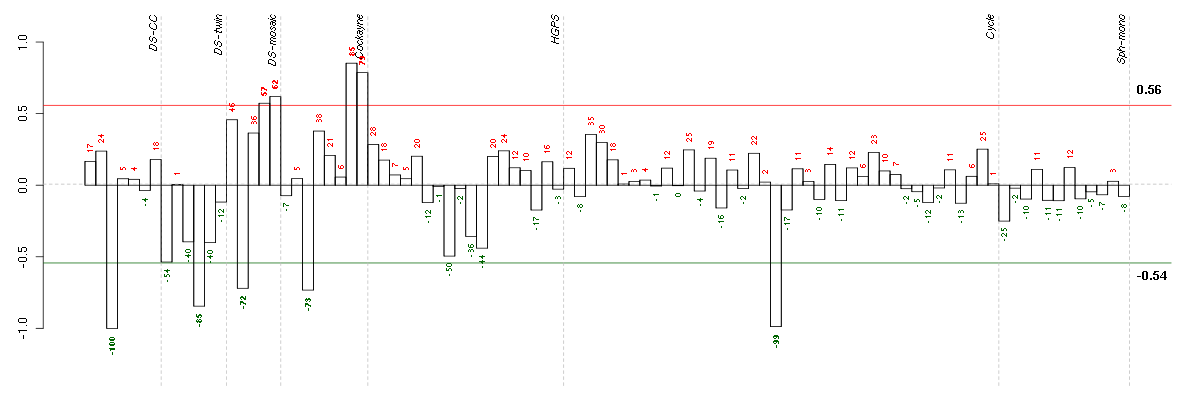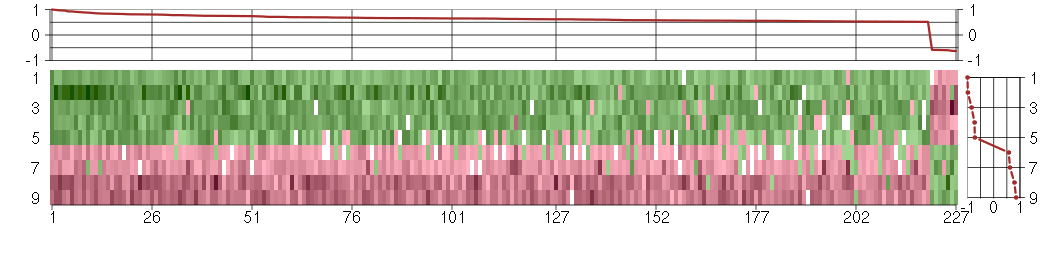

Under-expression is coded with green,
over-expression with red color.

vesicle-mediated transport
The directed movement of substances, either within a vesicle or in the vesicle membrane, into, out of or within a cell.
transport
The directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells, or within a multicellular organism.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
intracellular transport
The directed movement of substances within a cell.
localization
Any process by which a cell, a substance, or a cellular entity, such as a protein complex or organelle, is transported to, and/or maintained in a specific location.
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.
cellular localization
Any process by which a substance or cellular entity, such as a protein complex or organelle, is transported to, and/or maintained in, a specific location within or in the membrane of a cell.
establishment of localization in cell
The directed movement of a substance or cellular entity, such as a protein complex or organelle, to a specific location within, or in the membrane of, a cell.
all
This term is the most general term possible
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.
cellular localization
Any process by which a substance or cellular entity, such as a protein complex or organelle, is transported to, and/or maintained in, a specific location within or in the membrane of a cell.
establishment of localization in cell
The directed movement of a substance or cellular entity, such as a protein complex or organelle, to a specific location within, or in the membrane of, a cell.
vesicle-mediated transport
The directed movement of substances, either within a vesicle or in the vesicle membrane, into, out of or within a cell.
intracellular transport
The directed movement of substances within a cell.

intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
cytoplasm
All of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
cytoplasmic membrane-bounded vesicle
A membrane-bounded vesicle found in the cytoplasm of the cell.
cytoplasmic vesicle
A vesicle formed of membrane or protein, found in the cytoplasm of a cell.
vesicle
Any small, fluid-filled, spherical organelle enclosed by membrane or protein.
membrane-bounded vesicle
Any small, fluid-filled, spherical organelle enclosed by a lipid bilayer.
melanosome
A tissue-specific, membrane-bounded cytoplasmic organelle within which melanin pigments are synthesized and stored. Melanosomes are synthesized in melanocyte cells.
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
pigment granule
A small, subcellular membrane-bounded vesicle containing pigment and/or pigment precursor molecules. Pigment granule biogenesis is poorly understood, as pigment granules are derived from multiple sources including the endoplasmic reticulum, coated vesicles, lysosomes, and endosomes.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
cytoplasmic vesicle
A vesicle formed of membrane or protein, found in the cytoplasm of a cell.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
cytoplasmic membrane-bounded vesicle
A membrane-bounded vesicle found in the cytoplasm of the cell.
cytoplasmic membrane-bounded vesicle
A membrane-bounded vesicle found in the cytoplasm of the cell.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
cytoplasmic vesicle
A vesicle formed of membrane or protein, found in the cytoplasm of a cell.

protein binding
Interacting selectively with any protein or protein complex (a complex of two or more proteins that may include other nonprotein molecules).
molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
binding
The selective, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
all
This term is the most general term possible
| Id | Pvalue | ExpCount | Count | Size |
|---|---|---|---|---|
| miR-320/320abcd | 9.370e-03 | 12.81 | 32 | 372 |
ACLYATP citrate lyase (210337_s_at), score: 0.65 ACTR2ARP2 actin-related protein 2 homolog (yeast) (200727_s_at), score: 0.54 ADAM15ADAM metallopeptidase domain 15 (217007_s_at), score: 0.57 ADRBK1adrenergic, beta, receptor kinase 1 (201401_s_at), score: 0.58 AFF4AF4/FMR2 family, member 4 (219199_at), score: 0.6 AGFG1ArfGAP with FG repeats 1 (213926_s_at), score: 0.56 ANKFY1ankyrin repeat and FYVE domain containing 1 (219868_s_at), score: 0.63 AP1S1adaptor-related protein complex 1, sigma 1 subunit (205195_at), score: 0.58 ARF1ADP-ribosylation factor 1 (208750_s_at), score: 0.75 ARFGEF1ADP-ribosylation factor guanine nucleotide-exchange factor 1(brefeldin A-inhibited) (216266_s_at), score: 0.54 ARHGDIARho GDP dissociation inhibitor (GDI) alpha (213606_s_at), score: 0.75 ASPHaspartate beta-hydroxylase (205808_at), score: 0.62 ATP13A3ATPase type 13A3 (219558_at), score: 0.68 ATP6V1AATPase, H+ transporting, lysosomal 70kDa, V1 subunit A (201971_s_at), score: 0.81 BAG5BCL2-associated athanogene 5 (202984_s_at), score: 0.53 BCHEbutyrylcholinesterase (205433_at), score: 0.63 BCL2L1BCL2-like 1 (215037_s_at), score: 0.7 BET1Lblocked early in transport 1 homolog (S. cerevisiae)-like (220470_at), score: 0.67 BSGbasigin (Ok blood group) (208677_s_at), score: 0.65 C14orf118chromosome 14 open reading frame 118 (219720_s_at), score: 0.54 C19orf6chromosome 19 open reading frame 6 (213986_s_at), score: 0.88 C1orf144chromosome 1 open reading frame 144 (212003_at), score: 0.52 CACNA2D1calcium channel, voltage-dependent, alpha 2/delta subunit 1 (207050_at), score: 0.74 CALUcalumenin (214845_s_at), score: 0.74 CANXcalnexin (208853_s_at), score: 0.78 CAPRIN1cell cycle associated protein 1 (200722_s_at), score: 0.65 CBFBcore-binding factor, beta subunit (206788_s_at), score: 0.58 CD44CD44 molecule (Indian blood group) (210916_s_at), score: 0.82 CD46CD46 molecule, complement regulatory protein (211574_s_at), score: 0.56 CDK6cyclin-dependent kinase 6 (207143_at), score: 0.62 CDV3CDV3 homolog (mouse) (213548_s_at), score: 0.84 CHMchoroideremia (Rab escort protein 1) (207099_s_at), score: 0.61 CLIC4chloride intracellular channel 4 (201559_s_at), score: 0.65 CLINT1clathrin interactor 1 (201768_s_at), score: 0.56 CLIP1CAP-GLY domain containing linker protein 1 (210716_s_at), score: 0.52 CLPTM1cleft lip and palate associated transmembrane protein 1 (211136_s_at), score: 0.58 CLTCL1clathrin, heavy chain-like 1 (205944_s_at), score: -0.58 COL1A1collagen, type I, alpha 1 (217430_x_at), score: 0.73 COL6A1collagen, type VI, alpha 1 (212940_at), score: 0.84 COPAcoatomer protein complex, subunit alpha (214336_s_at), score: 0.98 CPDcarboxypeptidase D (201942_s_at), score: 0.85 CPNE3copine III (202118_s_at), score: 0.54 CSNK1A1casein kinase 1, alpha 1 (206562_s_at), score: 0.52 CYP51A1cytochrome P450, family 51, subfamily A, polypeptide 1 (216607_s_at), score: 0.58 CYTH3cytohesin 3 (206523_at), score: 0.67 DAPK3death-associated protein kinase 3 (203890_s_at), score: 0.81 DAZAP2DAZ associated protein 2 (212595_s_at), score: 0.56 DCBLD2discoidin, CUB and LCCL domain containing 2 (213865_at), score: 0.55 DCTN5dynactin 5 (p25) (209231_s_at), score: 0.54 DDX3XDEAD (Asp-Glu-Ala-Asp) box polypeptide 3, X-linked (201211_s_at), score: 0.92 DHX9DEAH (Asp-Glu-Ala-His) box polypeptide 9 (212105_s_at), score: 0.96 DNAJB1DnaJ (Hsp40) homolog, subfamily B, member 1 (200664_s_at), score: 0.59 DSTdystonin (212253_x_at), score: 0.69 DTX3deltex homolog 3 (Drosophila) (49049_at), score: 0.56 DUSP10dual specificity phosphatase 10 (215501_s_at), score: 0.61 EDIL3EGF-like repeats and discoidin I-like domains 3 (207379_at), score: 0.52 EHMT1euchromatic histone-lysine N-methyltransferase 1 (219339_s_at), score: 0.57 EIF4G1eukaryotic translation initiation factor 4 gamma, 1 (208624_s_at), score: 0.54 EIF5Aeukaryotic translation initiation factor 5A (201123_s_at), score: 0.65 ELK1ELK1, member of ETS oncogene family (210376_x_at), score: 0.74 ENSAendosulfine alpha (221487_s_at), score: 0.63 EPB41L2erythrocyte membrane protein band 4.1-like 2 (201718_s_at), score: 0.58 EPRSglutamyl-prolyl-tRNA synthetase (200841_s_at), score: 0.66 ERLIN1ER lipid raft associated 1 (202444_s_at), score: 0.73 ETS1v-ets erythroblastosis virus E26 oncogene homolog 1 (avian) (214447_at), score: 0.58 EXOC5exocyst complex component 5 (218748_s_at), score: 0.66 EZRezrin (208621_s_at), score: 0.62 FAM108B1family with sequence similarity 108, member B1 (220285_at), score: 0.55 FBN1fibrillin 1 (202765_s_at), score: 0.58 FGFR1fibroblast growth factor receptor 1 (210973_s_at), score: 0.7 FKBP15FK506 binding protein 15, 133kDa (76897_s_at), score: 0.6 FKBP1AFK506 binding protein 1A, 12kDa (210186_s_at), score: 0.63 FN1fibronectin 1 (214701_s_at), score: 0.9 FNTBfarnesyltransferase, CAAX box, beta (204764_at), score: 0.62 FUSfusion (involved in t(12;16) in malignant liposarcoma) (217370_x_at), score: 0.59 GABPAGA binding protein transcription factor, alpha subunit 60kDa (210188_at), score: 0.57 GALNT2UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-T2) (217787_s_at), score: 0.57 GDE1glycerophosphodiester phosphodiesterase 1 (202593_s_at), score: 0.53 GDI2GDP dissociation inhibitor 2 (200008_s_at), score: 0.62 GLULglutamate-ammonia ligase (glutamine synthetase) (200648_s_at), score: 0.71 GNA12guanine nucleotide binding protein (G protein) alpha 12 (221737_at), score: -0.6 GNA13guanine nucleotide binding protein (G protein), alpha 13 (206917_at), score: 0.66 GNSglucosamine (N-acetyl)-6-sulfatase (203676_at), score: 0.76 GPR137G protein-coupled receptor 137 (43934_at), score: 0.81 GPR176G protein-coupled receptor 176 (206673_at), score: 0.56 GTF2Igeneral transcription factor II, i (210892_s_at), score: 0.88 H2AFYH2A histone family, member Y (214500_at), score: 0.56 HIPK3homeodomain interacting protein kinase 3 (210148_at), score: 0.56 HMGA1high mobility group AT-hook 1 (210457_x_at), score: 0.65 HNRNPCheterogeneous nuclear ribonucleoprotein C (C1/C2) (200751_s_at), score: 0.6 HNRNPH1heterogeneous nuclear ribonucleoprotein H1 (H) (213470_s_at), score: 0.52 HP1BP3heterochromatin protein 1, binding protein 3 (220633_s_at), score: 0.8 HSPA4heat shock 70kDa protein 4 (211016_x_at), score: 0.67 IL13RA1interleukin 13 receptor, alpha 1 (210904_s_at), score: 0.53 INHBEinhibin, beta E (210587_at), score: -0.62 IPO8importin 8 (205701_at), score: 0.65 ITGA4integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor) (205885_s_at), score: 0.52 KCTD20potassium channel tetramerisation domain containing 20 (214849_at), score: 0.71 KDELR1KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1 (200922_at), score: 0.53 KPNA4karyopherin alpha 4 (importin alpha 3) (209653_at), score: 0.7 LAMP1lysosomal-associated membrane protein 1 (201551_s_at), score: 0.81 LATS1LATS, large tumor suppressor, homolog 1 (Drosophila) (219813_at), score: 0.53 LMAN1lectin, mannose-binding, 1 (203294_s_at), score: 0.68 LOC285412similar to KIAA0737 protein (217448_s_at), score: 0.68 LOC729148nuclear undecaprenyl pyrophosphate synthase 1 pseudogene (215207_x_at), score: 0.57 LOC730092RRN3 RNA polymerase I transcription factor homolog (S. cerevisiae) pseudogene (216908_x_at), score: 0.77 LOXlysyl oxidase (213640_s_at), score: 0.56 LOXL2lysyl oxidase-like 2 (202997_s_at), score: 0.64 LSSlanosterol synthase (2,3-oxidosqualene-lanosterol cyclase) (211019_s_at), score: 0.68 M6PRmannose-6-phosphate receptor (cation dependent) (200900_s_at), score: 0.57 MADDMAP-kinase activating death domain (38398_at), score: -0.63 MAP3K2mitogen-activated protein kinase kinase kinase 2 (221695_s_at), score: 0.62 MAP3K7mitogen-activated protein kinase kinase kinase 7 (211537_x_at), score: 0.65 MAP3K7IP2mitogen-activated protein kinase kinase kinase 7 interacting protein 2 (210284_s_at), score: 0.56 MAP4K5mitogen-activated protein kinase kinase kinase kinase 5 (203553_s_at), score: 0.57 MAPK1mitogen-activated protein kinase 1 (208351_s_at), score: 0.69 MAPKAPK2mitogen-activated protein kinase-activated protein kinase 2 (201461_s_at), score: 0.75 MAT2Amethionine adenosyltransferase II, alpha (200769_s_at), score: 0.83 MAXMYC associated factor X (210734_x_at), score: 0.91 MAZMYC-associated zinc finger protein (purine-binding transcription factor) (207824_s_at), score: 0.75 MBNL2muscleblind-like 2 (Drosophila) (205018_s_at), score: 0.66 MBTPS2membrane-bound transcription factor peptidase, site 2 (206473_at), score: 0.61 METmet proto-oncogene (hepatocyte growth factor receptor) (211599_x_at), score: 0.55 MFN2mitofusin 2 (216205_s_at), score: 0.8 MLXMAX-like protein X (217909_s_at), score: 0.53 MMP14matrix metallopeptidase 14 (membrane-inserted) (202827_s_at), score: 0.62 MOBKL1BMOB1, Mps One Binder kinase activator-like 1B (yeast) (201299_s_at), score: 0.54 MPDU1mannose-P-dolichol utilization defect 1 (209208_at), score: 0.68 NAB2NGFI-A binding protein 2 (EGR1 binding protein 2) (216017_s_at), score: 0.57 NBPF14neuroblastoma breakpoint family, member 14 (201104_x_at), score: 0.52 NDRG3NDRG family member 3 (221082_s_at), score: 0.56 NONOnon-POU domain containing, octamer-binding (208698_s_at), score: 0.65 NOTCH2Notch homolog 2 (Drosophila) (210756_s_at), score: 0.7 NRBP1nuclear receptor binding protein 1 (217765_at), score: 0.62 NUMA1nuclear mitotic apparatus protein 1 (214251_s_at), score: 0.75 PAFAH1B1platelet-activating factor acetylhydrolase, isoform Ib, alpha subunit 45kDa (211547_s_at), score: 0.8 PCDHGA11protocadherin gamma subfamily A, 11 (211876_x_at), score: 0.93 PCDHGA3protocadherin gamma subfamily A, 3 (216352_x_at), score: 0.96 PCIF1PDX1 C-terminal inhibiting factor 1 (222045_s_at), score: 0.57 PEA15phosphoprotein enriched in astrocytes 15 (200787_s_at), score: 0.52 PHTF2putative homeodomain transcription factor 2 (217097_s_at), score: 0.78 PICALMphosphatidylinositol binding clathrin assembly protein (215236_s_at), score: 0.74 PIP5K1Aphosphatidylinositol-4-phosphate 5-kinase, type I, alpha (211205_x_at), score: 0.8 PLEC1plectin 1, intermediate filament binding protein 500kDa (216971_s_at), score: 0.53 POLDIP3polymerase (DNA-directed), delta interacting protein 3 (215357_s_at), score: 0.81 POLR2Epolymerase (RNA) II (DNA directed) polypeptide E, 25kDa (213887_s_at), score: 0.56 POM121POM121 membrane glycoprotein (rat) (212178_s_at), score: 0.56 PPP2R1Aprotein phosphatase 2 (formerly 2A), regulatory subunit A, alpha isoform (200695_at), score: 0.59 PPP2R5Dprotein phosphatase 2, regulatory subunit B', delta isoform (211159_s_at), score: 0.66 PPP5Cprotein phosphatase 5, catalytic subunit (201979_s_at), score: 0.53 PQLC1PQ loop repeat containing 1 (218208_at), score: -0.59 PRKAB1protein kinase, AMP-activated, beta 1 non-catalytic subunit (201835_s_at), score: 0.66 PRKAR1Aprotein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1) (200604_s_at), score: 0.65 PRPF4PRP4 pre-mRNA processing factor 4 homolog (yeast) (209162_s_at), score: 0.54 PRPF4BPRP4 pre-mRNA processing factor 4 homolog B (yeast) (211090_s_at), score: 0.62 PRPF6PRP6 pre-mRNA processing factor 6 homolog (S. cerevisiae) (208879_x_at), score: 0.64 PRRX1paired related homeobox 1 (205991_s_at), score: 0.75 PSEN1presenilin 1 (207782_s_at), score: 0.63 PSME4proteasome (prosome, macropain) activator subunit 4 (212220_at), score: 0.58 PTBP1polypyrimidine tract binding protein 1 (212016_s_at), score: 1 PTMSparathymosin (218045_x_at), score: 0.51 PTPN11protein tyrosine phosphatase, non-receptor type 11 (209896_s_at), score: 0.55 PTPN14protein tyrosine phosphatase, non-receptor type 14 (205503_at), score: 0.71 PTPRAprotein tyrosine phosphatase, receptor type, A (213799_s_at), score: 0.63 PTPROprotein tyrosine phosphatase, receptor type, O (211600_at), score: 0.56 PTRFpolymerase I and transcript release factor (208790_s_at), score: 0.56 PXNpaxillin (211823_s_at), score: 0.84 RAB11BRAB11B, member RAS oncogene family (34478_at), score: 0.52 RAB35RAB35, member RAS oncogene family (205461_at), score: 0.67 RAB5ARAB5A, member RAS oncogene family (206113_s_at), score: 0.87 RAB5CRAB5C, member RAS oncogene family (201156_s_at), score: 0.78 RANBP2RAN binding protein 2 (201711_x_at), score: 0.56 RASA3RAS p21 protein activator 3 (206220_s_at), score: 0.66 RBM9RNA binding motif protein 9 (213901_x_at), score: 0.52 RCAN1regulator of calcineurin 1 (215253_s_at), score: 0.52 RELAv-rel reticuloendotheliosis viral oncogene homolog A (avian) (209878_s_at), score: 0.66 RRN3RRN3 RNA polymerase I transcription factor homolog (S. cerevisiae) (216902_s_at), score: 0.69 RRP7Bribosomal RNA processing 7 homolog B (S. cerevisiae) (214828_s_at), score: 0.66 RXRBretinoid X receptor, beta (215099_s_at), score: 0.83 SCAMP1secretory carrier membrane protein 1 (206667_s_at), score: 0.68 SCARB2scavenger receptor class B, member 2 (201647_s_at), score: 0.65 SDHCsuccinate dehydrogenase complex, subunit C, integral membrane protein, 15kDa (216591_s_at), score: 0.61 SENP3SUMO1/sentrin/SMT3 specific peptidase 3 (215113_s_at), score: 0.7 SEPT11septin 11 (201308_s_at), score: 0.71 SEPT2septin 2 (200778_s_at), score: 0.56 SFRS1splicing factor, arginine/serine-rich 1 (201742_x_at), score: 0.65 SH3GLB2SH3-domain GRB2-like endophilin B2 (218813_s_at), score: 0.56 SHC1SHC (Src homology 2 domain containing) transforming protein 1 (201469_s_at), score: 0.66 SLC12A4solute carrier family 12 (potassium/chloride transporters), member 4 (209401_s_at), score: 0.72 SLC6A9solute carrier family 6 (neurotransmitter transporter, glycine), member 9 (207043_s_at), score: 0.65 SMARCA2SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 (212257_s_at), score: 0.52 SMEK1SMEK homolog 1, suppressor of mek1 (Dictyostelium) (220368_s_at), score: 0.75 SNAP23synaptosomal-associated protein, 23kDa (214544_s_at), score: 0.7 SPAG9sperm associated antigen 9 (206748_s_at), score: 0.67 SPPL2Bsignal peptide peptidase-like 2B (215833_s_at), score: 0.66 SPSB3splA/ryanodine receptor domain and SOCS box containing 3 (46256_at), score: -0.58 SRPRsignal recognition particle receptor (docking protein) (200917_s_at), score: 0.76 SS18synovial sarcoma translocation, chromosome 18 (216684_s_at), score: 0.69 STAT2signal transducer and activator of transcription 2, 113kDa (205170_at), score: 0.6 STIP1stress-induced-phosphoprotein 1 (212009_s_at), score: 0.79 SYPL1synaptophysin-like 1 (201259_s_at), score: 0.78 TAF9BTAF9B RNA polymerase II, TATA box binding protein (TBP)-associated factor, 31kDa (221618_s_at), score: 0.61 TGFBR2transforming growth factor, beta receptor II (70/80kDa) (207334_s_at), score: 0.83 TJP1tight junction protein 1 (zona occludens 1) (214168_s_at), score: 0.58 TM9SF4transmembrane 9 superfamily protein member 4 (212194_s_at), score: 0.68 TMCO3transmembrane and coiled-coil domains 3 (220240_s_at), score: 0.64 TMED2transmembrane emp24 domain trafficking protein 2 (204426_at), score: 0.71 TMOD3tropomodulin 3 (ubiquitous) (220800_s_at), score: 0.63 TMX1thioredoxin-related transmembrane protein 1 (208097_s_at), score: 0.55 TOM1L2target of myb1-like 2 (chicken) (214840_at), score: 0.54 TOR1AIP1torsin A interacting protein 1 (216100_s_at), score: 0.58 TRIM25tripartite motif-containing 25 (206911_at), score: 0.57 TUBB2Atubulin, beta 2A (209372_x_at), score: 0.62 TWF1twinfilin, actin-binding protein, homolog 1 (Drosophila) (214007_s_at), score: 0.53 UBE2D3ubiquitin-conjugating enzyme E2D 3 (UBC4/5 homolog, yeast) (200669_s_at), score: 0.66 UBXN4UBX domain protein 4 (212008_at), score: 0.64 UTRNutrophin (213022_s_at), score: 0.74 VAMP3vesicle-associated membrane protein 3 (cellubrevin) (201337_s_at), score: 0.69 VCANversican (215646_s_at), score: 0.53 WACWW domain containing adaptor with coiled-coil (219679_s_at), score: 0.81 WDR1WD repeat domain 1 (210935_s_at), score: 0.77 YIPF5Yip1 domain family, member 5 (221423_s_at), score: 0.65 YWHAEtyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide (210317_s_at), score: 0.75 YWHAZtyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide (200641_s_at), score: 0.68 ZFAND5zinc finger, AN1-type domain 5 (217741_s_at), score: 0.62 ZFP36L2zinc finger protein 36, C3H type-like 2 (201367_s_at), score: 0.52 ZNF26zinc finger protein 26 (219595_at), score: -0.59
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| ctrl c 08-03.CEL | 3 | 1 | DS-CC | hgu133a | none | DS-CC 3 |
| E-TABM-263-raw-cel-1515486031.cel | 20 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 4Twin.CEL | 4 | 2 | DS-twin | hgu133plus2 | none | DS-twin 4 |
| E-GEOD-3407-raw-cel-1437949655.cel | 3 | 4 | Cockayne | hgu133a | none | CSB |
| 46B.CEL | 2 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 2 |
| 47B.CEL | 4 | 3 | DS-mosaic | hgu133plus2 | Down mosaic | DS-mosaic 4 |
| 47C.CEL | 5 | 3 | DS-mosaic | hgu133plus2 | Down mosaic | DS-mosaic 5 |
| E-GEOD-3407-raw-cel-1437949938.cel | 8 | 4 | Cockayne | hgu133a | none | CSB |
| E-GEOD-3407-raw-cel-1437949854.cel | 7 | 4 | Cockayne | hgu133a | CS | eGFP |