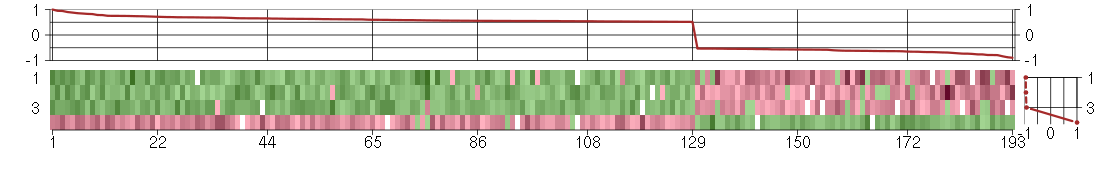

Under-expression is coded with green,
over-expression with red color.


plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
integral to membrane
Penetrating at least one phospholipid bilayer of a membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer. When used to describe a protein, indicates that all or part of the peptide sequence is embedded in the membrane.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.

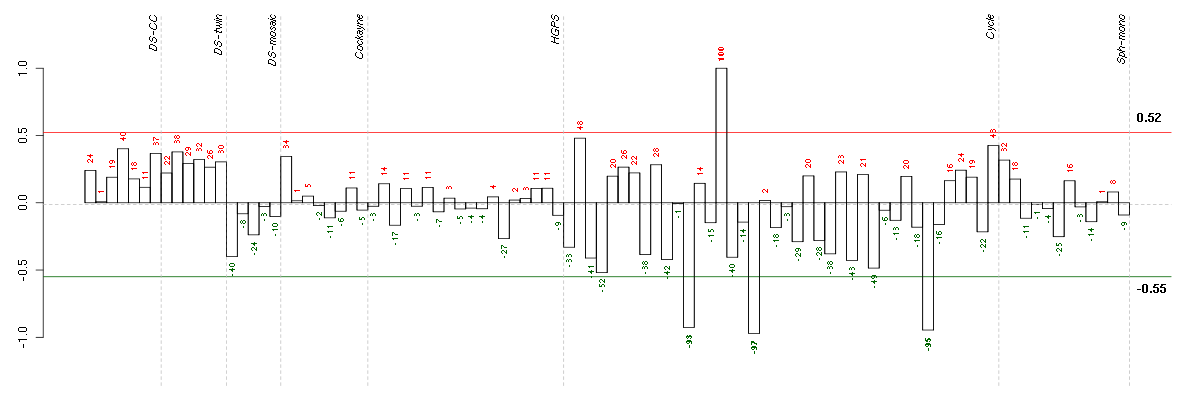
ACOT11acyl-CoA thioesterase 11 (216103_at), score: 0.55 ACSL5acyl-CoA synthetase long-chain family member 5 (218322_s_at), score: 0.6 ADORA1adenosine A1 receptor (216220_s_at), score: 0.63 AGMATagmatine ureohydrolase (agmatinase) (219792_at), score: 0.52 AHCYL2S-adenosylhomocysteine hydrolase-like 2 (212814_at), score: -0.67 ALDH1L1aldehyde dehydrogenase 1 family, member L1 (215798_at), score: 0.69 ALPPL2alkaline phosphatase, placental-like 2 (216377_x_at), score: 0.55 ANGEL1angel homolog 1 (Drosophila) (36865_at), score: 0.55 APOC4apolipoprotein C-IV (206738_at), score: 0.69 ART1ADP-ribosyltransferase 1 (207919_at), score: 0.56 ATF7IP2activating transcription factor 7 interacting protein 2 (219870_at), score: 0.53 ATP6V0A1ATPase, H+ transporting, lysosomal V0 subunit a1 (205095_s_at), score: -0.57 B4GALT1UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1 (201883_s_at), score: 0.58 BAI2brain-specific angiogenesis inhibitor 2 (204966_at), score: 0.6 BCL2L11BCL2-like 11 (apoptosis facilitator) (222343_at), score: 0.9 BCL3B-cell CLL/lymphoma 3 (204908_s_at), score: -0.63 BHLHE40basic helix-loop-helix family, member e40 (201170_s_at), score: -0.55 BIRC3baculoviral IAP repeat-containing 3 (210538_s_at), score: -0.7 C19orf40chromosome 19 open reading frame 40 (214816_x_at), score: 0.69 C1orf159chromosome 1 open reading frame 159 (219337_at), score: -0.63 C1orf89chromosome 1 open reading frame 89 (220963_s_at), score: 0.53 C6orf155chromosome 6 open reading frame 155 (220324_at), score: 0.52 C6orf62chromosome 6 open reading frame 62 (213875_x_at), score: 0.56 C9orf114chromosome 9 open reading frame 114 (218565_at), score: 0.65 CCDC40coiled-coil domain containing 40 (220592_at), score: 0.53 CCDC94coiled-coil domain containing 94 (204335_at), score: 0.52 CD79BCD79b molecule, immunoglobulin-associated beta (205297_s_at), score: 0.86 CDH19cadherin 19, type 2 (206898_at), score: 0.55 CEACAM1carcinoembryonic antigen-related cell adhesion molecule 1 (biliary glycoprotein) (211883_x_at), score: 0.64 CELSR2cadherin, EGF LAG seven-pass G-type receptor 2 (flamingo homolog, Drosophila) (204029_at), score: 0.57 CFIcomplement factor I (203854_at), score: -0.69 CHRNA6cholinergic receptor, nicotinic, alpha 6 (207568_at), score: 0.64 CLCF1cardiotrophin-like cytokine factor 1 (219500_at), score: -0.62 CLEC1AC-type lectin domain family 1, member A (219761_at), score: -0.73 CNNM3cyclin M3 (220739_s_at), score: -0.64 COL11A2collagen, type XI, alpha 2 (216993_s_at), score: 0.58 CR1complement component (3b/4b) receptor 1 (Knops blood group) (217484_at), score: 0.52 CRYBB2crystallin, beta B2 (206777_s_at), score: 0.53 CRYL1crystallin, lambda 1 (220753_s_at), score: -0.56 CTTNcortactin (201059_at), score: 0.56 CXCL11chemokine (C-X-C motif) ligand 11 (210163_at), score: -0.57 CXorf21chromosome X open reading frame 21 (220252_x_at), score: 0.68 CYP24A1cytochrome P450, family 24, subfamily A, polypeptide 1 (206504_at), score: 0.64 CYP3A4cytochrome P450, family 3, subfamily A, polypeptide 4 (205998_x_at), score: 0.62 CYP7B1cytochrome P450, family 7, subfamily B, polypeptide 1 (207386_at), score: -0.72 DKFZp547G183hypothetical LOC55525 (220572_at), score: 0.66 DNAJC2DnaJ (Hsp40) homolog, subfamily C, member 2 (213097_s_at), score: 0.54 DNM3dynamin 3 (209839_at), score: -0.62 DSPPdentin sialophosphoprotein (221681_s_at), score: 0.57 DUSP5dual specificity phosphatase 5 (209457_at), score: -0.67 EDNRBendothelin receptor type B (204271_s_at), score: -0.63 EGR2early growth response 2 (Krox-20 homolog, Drosophila) (205249_at), score: -0.54 EGR3early growth response 3 (206115_at), score: -0.78 EHMT2euchromatic histone-lysine N-methyltransferase 2 (202326_at), score: -0.58 ELSPBP1epididymal sperm binding protein 1 (220366_at), score: 0.6 ENTPD1ectonucleoside triphosphate diphosphohydrolase 1 (209473_at), score: 0.53 EPHA2EPH receptor A2 (203499_at), score: -0.57 ERFEts2 repressor factor (203643_at), score: 0.56 F11coagulation factor XI (206610_s_at), score: 0.61 FBXO2F-box protein 2 (219305_x_at), score: 0.59 FETUBfetuin B (214417_s_at), score: 0.7 FKBP10FK506 binding protein 10, 65 kDa (219249_s_at), score: 0.74 FUT2fucosyltransferase 2 (secretor status included) (210608_s_at), score: 0.63 GABRG3gamma-aminobutyric acid (GABA) A receptor, gamma 3 (216895_at), score: 0.65 GASTgastrin (208138_at), score: 0.55 GFOD1glucose-fructose oxidoreductase domain containing 1 (219821_s_at), score: 0.59 GLRA1glycine receptor, alpha 1 (207972_at), score: 0.52 GPATCH4G patch domain containing 4 (220596_at), score: 0.95 GPX3glutathione peroxidase 3 (plasma) (214091_s_at), score: -0.56 GZMMgranzyme M (lymphocyte met-ase 1) (207460_at), score: 0.55 HAB1B1 for mucin (215778_x_at), score: 0.62 HAND1heart and neural crest derivatives expressed 1 (220138_at), score: 0.69 HES1hairy and enhancer of split 1, (Drosophila) (203394_s_at), score: -0.76 HIST1H2BGhistone cluster 1, H2bg (210387_at), score: -0.66 HMGA1high mobility group AT-hook 1 (210457_x_at), score: -0.54 HNRNPH1heterogeneous nuclear ribonucleoprotein H1 (H) (213470_s_at), score: 0.53 HPNhepsin (204934_s_at), score: 0.7 IER2immediate early response 2 (202081_at), score: -0.66 IGHG1immunoglobulin heavy constant gamma 1 (G1m marker) (211693_at), score: 0.64 IL2RBinterleukin 2 receptor, beta (205291_at), score: 0.83 IL9Rinterleukin 9 receptor (217212_s_at), score: 1 INAinternexin neuronal intermediate filament protein, alpha (204465_s_at), score: -0.65 INE1inactivation escape 1 (non-protein coding) (207252_at), score: 0.56 INHAinhibin, alpha (210141_s_at), score: 0.68 ITGALintegrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide) (213475_s_at), score: 0.72 JAG2jagged 2 (32137_at), score: 0.7 JAK2Janus kinase 2 (a protein tyrosine kinase) (205842_s_at), score: -0.68 JUNBjun B proto-oncogene (201473_at), score: -0.87 JUPjunction plakoglobin (201015_s_at), score: -0.58 KIAA1009KIAA1009 (206005_s_at), score: 0.62 KLHL5kelch-like 5 (Drosophila) (220682_s_at), score: 0.69 KRI1KRI1 homolog (S. cerevisiae) (218798_at), score: 0.55 LAX1lymphocyte transmembrane adaptor 1 (207734_at), score: 0.63 LOC100188945cell division cycle associated 4 pseudogene (215109_at), score: 0.61 LOC388152hypothetical LOC388152 (220602_s_at), score: 0.58 LOC391132similar to hCG2041276 (216177_at), score: 0.75 LOC80054hypothetical LOC80054 (220465_at), score: 0.54 LRRC37B2leucine rich repeat containing 37, member B2 (216149_at), score: 0.55 LSM14BLSM14B, SCD6 homolog B (S. cerevisiae) (219653_at), score: 0.52 MAFKv-maf musculoaponeurotic fibrosarcoma oncogene homolog K (avian) (206750_at), score: 0.55 MAP3K14mitogen-activated protein kinase kinase kinase 14 (205192_at), score: -0.57 MFNGMFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase (204153_s_at), score: 0.54 MGC5590hypothetical protein MGC5590 (220931_at), score: 0.55 MLYCDmalonyl-CoA decarboxylase (218869_at), score: -0.54 MUC1mucin 1, cell surface associated (207847_s_at), score: -0.79 MUC5ACmucin 5AC, oligomeric mucus/gel-forming (217182_at), score: 0.65 MYH15myosin, heavy chain 15 (215331_at), score: 0.88 MYL10myosin, light chain 10, regulatory (221659_s_at), score: 0.67 MYO15Amyosin XVA (220288_at), score: 0.71 NACA2nascent polypeptide-associated complex alpha subunit 2 (222224_at), score: 0.53 NCOA2nuclear receptor coactivator 2 (205732_s_at), score: -0.63 NDRG3NDRG family member 3 (221082_s_at), score: -0.54 NFKBIAnuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha (201502_s_at), score: -0.63 NOVA1neuro-oncological ventral antigen 1 (205794_s_at), score: 0.54 NPAL2NIPA-like domain containing 2 (220128_s_at), score: -0.63 NSUN5BNOL1/NOP2/Sun domain family, member 5B (214100_x_at), score: 0.56 OAS22'-5'-oligoadenylate synthetase 2, 69/71kDa (204972_at), score: -0.53 OCLMoculomedin (208274_at), score: 0.74 OLAHoleoyl-ACP hydrolase (219975_x_at), score: 0.73 OR7E47Polfactory receptor, family 7, subfamily E, member 47 pseudogene (216698_x_at), score: 0.56 PACS1phosphofurin acidic cluster sorting protein 1 (220557_s_at), score: -0.83 PAPPA2pappalysin 2 (213332_at), score: 0.71 PCDHGA3protocadherin gamma subfamily A, 3 (216352_x_at), score: -0.57 PCOTHprostate collagen triple helix (222277_at), score: 0.55 PGLYRP4peptidoglycan recognition protein 4 (220944_at), score: 0.56 PIPOXpipecolic acid oxidase (221605_s_at), score: 0.84 PLA2G6phospholipase A2, group VI (cytosolic, calcium-independent) (215938_s_at), score: 0.73 PLA2R1phospholipase A2 receptor 1, 180kDa (207415_at), score: 0.53 POM121L2POM121 membrane glycoprotein-like 2 (rat) (216582_at), score: 0.55 PRKCBprotein kinase C, beta (209685_s_at), score: 0.82 PRKYprotein kinase, Y-linked (206279_at), score: -0.66 PTPN6protein tyrosine phosphatase, non-receptor type 6 (206687_s_at), score: 0.55 PVT1Pvt1 oncogene (non-protein coding) (216249_at), score: 0.66 PXNpaxillin (211823_s_at), score: -0.6 RAP1GAPRAP1 GTPase activating protein (203911_at), score: 0.75 RASGRP3RAS guanyl releasing protein 3 (calcium and DAG-regulated) (205801_s_at), score: -0.74 RBM38RNA binding motif protein 38 (212430_at), score: 0.55 RIPK1receptor (TNFRSF)-interacting serine-threonine kinase 1 (209941_at), score: -0.55 RLN1relaxin 1 (211753_s_at), score: 0.94 ROS1c-ros oncogene 1 , receptor tyrosine kinase (207569_at), score: 0.53 RPGRIP1retinitis pigmentosa GTPase regulator interacting protein 1 (206608_s_at), score: 0.65 RPL10Lribosomal protein L10-like (217559_at), score: 0.56 RPL27Aribosomal protein L27a (212044_s_at), score: 0.68 RPLP2ribosomal protein, large, P2 (200908_s_at), score: 0.52 RPS11ribosomal protein S11 (213350_at), score: 0.52 RXRBretinoid X receptor, beta (215099_s_at), score: -0.57 S100A14S100 calcium binding protein A14 (218677_at), score: 0.64 SAA4serum amyloid A4, constitutive (207096_at), score: 0.54 SARM1sterile alpha and TIR motif containing 1 (213259_s_at), score: 0.58 SDPRserum deprivation response (phosphatidylserine binding protein) (218711_s_at), score: -0.61 SEPT5septin 5 (209767_s_at), score: 0.62 SIGLEC1sialic acid binding Ig-like lectin 1, sialoadhesin (44673_at), score: 0.61 SIN3BSIN3 homolog B, transcription regulator (yeast) (209352_s_at), score: -0.53 SLC14A1solute carrier family 14 (urea transporter), member 1 (Kidd blood group) (205856_at), score: -0.55 SLC15A1solute carrier family 15 (oligopeptide transporter), member 1 (211349_at), score: 0.58 SLC2A8solute carrier family 2 (facilitated glucose transporter), member 8 (218985_at), score: -0.57 SLFN12schlafen family member 12 (219885_at), score: -0.56 SMAD7SMAD family member 7 (204790_at), score: -0.55 SNAI1snail homolog 1 (Drosophila) (219480_at), score: -0.58 SNAP25synaptosomal-associated protein, 25kDa (202508_s_at), score: -0.89 SORBS1sorbin and SH3 domain containing 1 (218087_s_at), score: 0.63 SPAG8sperm associated antigen 8 (206816_s_at), score: 0.59 ST8SIA3ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 (208064_s_at), score: 0.75 STIM1stromal interaction molecule 1 (202764_at), score: -0.53 SYT1synaptotagmin I (203999_at), score: -0.68 TAL1T-cell acute lymphocytic leukemia 1 (206283_s_at), score: 0.78 TBX10T-box 10 (207689_at), score: 0.54 TCP10t-complex 10 homolog (mouse) (207503_at), score: 0.61 TIAF1TGFB1-induced anti-apoptotic factor 1 (202039_at), score: -0.79 TLE3transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) (206472_s_at), score: -0.54 TMCC2transmembrane and coiled-coil domain family 2 (213096_at), score: 0.65 TMSB4Ythymosin beta 4, Y-linked (206769_at), score: 0.75 TNFSF12tumor necrosis factor (ligand) superfamily, member 12 (205611_at), score: -0.54 TNK1tyrosine kinase, non-receptor, 1 (217149_x_at), score: 0.69 TRA@T cell receptor alpha locus (216540_at), score: 0.63 TRAFD1TRAF-type zinc finger domain containing 1 (35254_at), score: -0.76 TRIB1tribbles homolog 1 (Drosophila) (202241_at), score: -0.73 TRIM45tripartite motif-containing 45 (219923_at), score: 0.55 TULP2tubby like protein 2 (206733_at), score: 0.78 TXNL4Bthioredoxin-like 4B (218794_s_at), score: 0.57 UGT1A8UDP glucuronosyltransferase 1 family, polypeptide A8 (221304_at), score: 0.54 VEGFBvascular endothelial growth factor B (203683_s_at), score: -0.53 WDR42AWD repeat domain 42A (202249_s_at), score: -0.61 ZC3H12Azinc finger CCCH-type containing 12A (218810_at), score: -0.62 ZKSCAN4zinc finger with KRAB and SCAN domains 4 (213625_at), score: 0.52 ZNF117zinc finger protein 117 (207605_x_at), score: 0.75 ZNF132zinc finger protein 132 (207402_at), score: -0.62 ZNF224zinc finger protein 224 (216983_s_at), score: 0.62 ZNF37Azinc finger protein 37A (214878_at), score: -0.62 ZNF394zinc finger protein 394 (214714_at), score: 0.52 ZNF701zinc finger protein 701 (220242_x_at), score: 0.51 ZNF783zinc finger family member 783 (221876_at), score: 0.59
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485991.cel | 18 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486311.cel | 34 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485931.cel | 15 | 6 | Cycle | hgu133a2 | none | Cycle 1 |