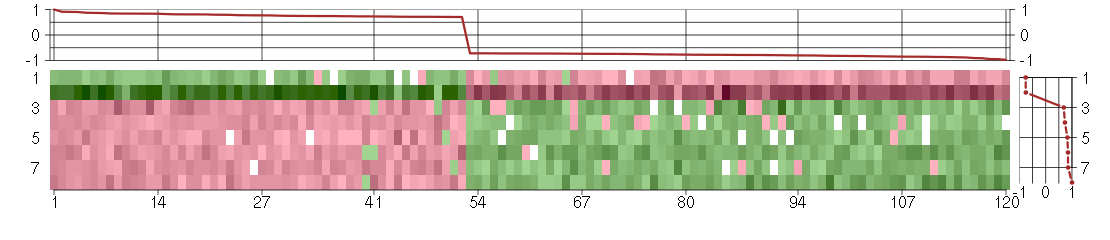

Under-expression is coded with green,
over-expression with red color.

biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
positive regulation of kinase activity
Any process that activates or increases the frequency, rate or extent of kinase activity, the catalysis of the transfer of a phosphate group, usually from ATP, to a substrate molecule.
positive regulation of catalytic activity
Any process that activates or increases the activity of an enzyme.
regulation of MAP kinase activity
Any process that modulates the frequency, rate or extent of MAP kinase activity.
positive regulation of MAP kinase activity
Any process that activates or increases the frequency, rate or extent of MAP kinase activity.
regulation of JUN kinase activity
Any process that modulates the frequency, rate or extent of JUN kinase activity.
positive regulation of JUN kinase activity
Any process that activates or increases the frequency, rate or extent of JUN kinase activity.
regulation of kinase activity
Any process that modulates the frequency, rate or extent of kinase activity, the catalysis of the transfer of a phosphate group, usually from ATP, to a substrate molecule.
regulation of protein kinase activity
Any process that modulates the frequency, rate or extent of protein kinase activity.
positive regulation of protein kinase activity
Any process that activates or increases the frequency, rate or extent of protein kinase activity.
regulation of catalytic activity
Any process that modulates the activity of an enzyme.
regulation of transferase activity
Any process that modulates the frequency, rate or extent of transferase activity, the catalysis of the transfer of a group, e.g. a methyl group, glycosyl group, acyl group, phosphorus-containing, or other groups, from one compound (generally regarded as the donor) to another compound (generally regarded as the acceptor). Transferase is the systematic name for any enzyme of EC class 2.
positive regulation of transferase activity
Any process that activates or increases the frequency, rate or extent of transferase activity, the catalysis of the transfer of a group, e.g. a methyl group, glycosyl group, acyl group, phosphorus-containing, or other groups, from a donor compound to an acceptor.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
regulation of molecular function
Any process that modulates the frequency, rate or extent of molecular functions. Molecular functions are elemental biological activities occurring at the molecular level, such as catalysis or binding.
all
This term is the most general term possible
positive regulation of transferase activity
Any process that activates or increases the frequency, rate or extent of transferase activity, the catalysis of the transfer of a group, e.g. a methyl group, glycosyl group, acyl group, phosphorus-containing, or other groups, from a donor compound to an acceptor.
positive regulation of kinase activity
Any process that activates or increases the frequency, rate or extent of kinase activity, the catalysis of the transfer of a phosphate group, usually from ATP, to a substrate molecule.
positive regulation of protein kinase activity
Any process that activates or increases the frequency, rate or extent of protein kinase activity.
positive regulation of MAP kinase activity
Any process that activates or increases the frequency, rate or extent of MAP kinase activity.
positive regulation of JUN kinase activity
Any process that activates or increases the frequency, rate or extent of JUN kinase activity.


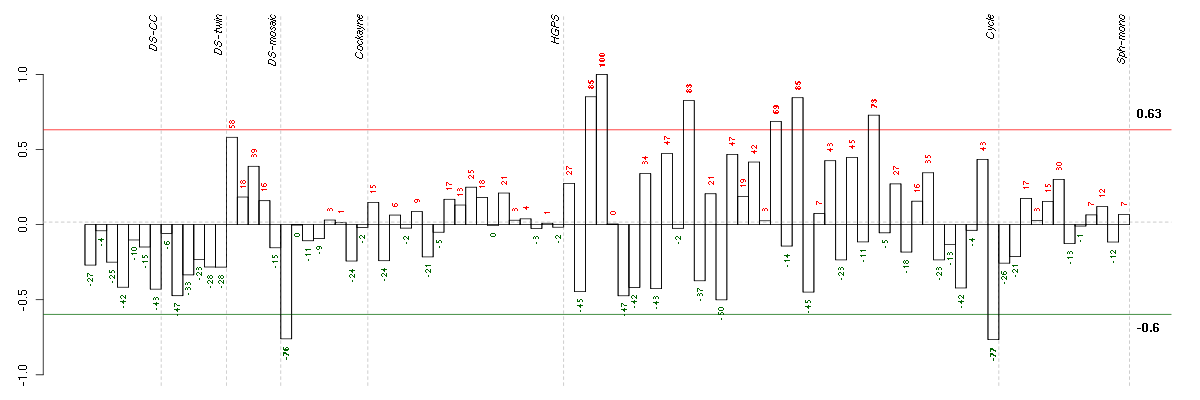
ADAMTS12ADAM metallopeptidase with thrombospondin type 1 motif, 12 (221421_s_at), score: -0.79 AIPL1aryl hydrocarbon receptor interacting protein-like 1 (219977_at), score: -0.8 ALDH1L1aldehyde dehydrogenase 1 family, member L1 (215798_at), score: -0.9 APOC4apolipoprotein C-IV (206738_at), score: -0.75 ARSAarylsulfatase A (204443_at), score: 0.78 BAP1BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) (201419_at), score: 0.75 BAT2HLA-B associated transcript 2 (212081_x_at), score: 0.87 BAT2LHLA-B associated transcript 2-like (212068_s_at), score: 0.85 BTNL3butyrophilin-like 3 (217207_s_at), score: -0.85 C1orf69chromosome 1 open reading frame 69 (215490_at), score: -0.75 CALB2calbindin 2 (205428_s_at), score: -0.79 CD79BCD79b molecule, immunoglobulin-associated beta (205297_s_at), score: -0.97 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (217849_s_at), score: 0.81 CHST4carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 4 (220446_s_at), score: -0.82 CICcapicua homolog (Drosophila) (212784_at), score: 0.77 COL11A2collagen, type XI, alpha 2 (216993_s_at), score: -0.86 CORO2Bcoronin, actin binding protein, 2B (209789_at), score: 0.72 COX6A2cytochrome c oxidase subunit VIa polypeptide 2 (206353_at), score: -0.82 CYP3A4cytochrome P450, family 3, subfamily A, polypeptide 4 (205998_x_at), score: -0.77 DKFZP564O0823DKFZP564O0823 protein (204687_at), score: -0.73 DNASE1L3deoxyribonuclease I-like 3 (205554_s_at), score: -0.81 DOK4docking protein 4 (209691_s_at), score: 0.71 EDARectodysplasin A receptor (220048_at), score: -0.82 EPAGearly lymphoid activation protein (217050_at), score: -0.72 ETV7ets variant 7 (221680_s_at), score: -0.85 EVLEnah/Vasp-like (217838_s_at), score: 0.72 FBP1fructose-1,6-bisphosphatase 1 (209696_at), score: -0.82 FCF1FCF1 small subunit (SSU) processome component homolog (S. cerevisiae) (219927_at), score: -0.73 FGL1fibrinogen-like 1 (205305_at), score: -0.73 FHOD1formin homology 2 domain containing 1 (218530_at), score: 0.74 FOXK2forkhead box K2 (203064_s_at), score: 0.75 FUT2fucosyltransferase 2 (secretor status included) (210608_s_at), score: -0.88 GABARAPL3GABA(A) receptors associated protein like 3 (pseudogene) (211458_s_at), score: -0.8 GPATCH4G patch domain containing 4 (220596_at), score: -0.72 GTPBP1GTP binding protein 1 (219357_at), score: 0.71 HAB1B1 for mucin (215778_x_at), score: -0.91 HAMPhepcidin antimicrobial peptide (220491_at), score: -0.84 HAO2hydroxyacid oxidase 2 (long chain) (220801_s_at), score: -0.8 HIST1H4Dhistone cluster 1, H4d (208076_at), score: -0.73 HS3ST2heparan sulfate (glucosamine) 3-O-sulfotransferase 2 (219697_at), score: -0.72 HSF1heat shock transcription factor 1 (202344_at), score: 0.76 HTR1F5-hydroxytryptamine (serotonin) receptor 1F (221458_at), score: -0.75 IDUAiduronidase, alpha-L- (205059_s_at), score: 0.91 IGHG1immunoglobulin heavy constant gamma 1 (G1m marker) (211693_at), score: -0.72 IL9Rinterleukin 9 receptor (217212_s_at), score: -0.84 INTS1integrator complex subunit 1 (212212_s_at), score: 0.77 ITGALintegrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide) (213475_s_at), score: -0.79 JUPjunction plakoglobin (201015_s_at), score: 0.87 LEPREL2leprecan-like 2 (204854_at), score: 0.84 LLGL2lethal giant larvae homolog 2 (Drosophila) (203713_s_at), score: -0.78 LOC149478hypothetical protein LOC149478 (215462_at), score: -0.77 LRDDleucine-rich repeats and death domain containing (221640_s_at), score: 0.84 LRP5low density lipoprotein receptor-related protein 5 (209468_at), score: 0.71 LRRN2leucine rich repeat neuronal 2 (216164_at), score: -0.79 MADCAM1mucosal vascular addressin cell adhesion molecule 1 (208037_s_at), score: -0.74 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: 0.9 MAP3K6mitogen-activated protein kinase kinase kinase 6 (219278_at), score: 1 MEOX1mesenchyme homeobox 1 (205619_s_at), score: -0.78 MOBPmyelin-associated oligodendrocyte basic protein (210193_at), score: -0.78 MUC5ACmucin 5AC, oligomeric mucus/gel-forming (217182_at), score: -0.74 MUL1mitochondrial E3 ubiquitin ligase 1 (218246_at), score: 0.74 MYH15myosin, heavy chain 15 (215331_at), score: -0.73 MYL4myosin, light chain 4, alkali; atrial, embryonic (210088_x_at), score: -0.76 NACAP1nascent-polypeptide-associated complex alpha polypeptide pseudogene 1 (211445_x_at), score: -0.74 NCRNA00093non-protein coding RNA 93 (210723_x_at), score: -0.74 NPDC1neural proliferation, differentiation and control, 1 (218086_at), score: 0.77 NTRK2neurotrophic tyrosine kinase, receptor, type 2 (207152_at), score: -0.72 NXPH3neurexophilin 3 (221991_at), score: -0.72 OR7E47Polfactory receptor, family 7, subfamily E, member 47 pseudogene (216698_x_at), score: -0.77 PAK1p21 protein (Cdc42/Rac)-activated kinase 1 (209615_s_at), score: 0.71 PARVBparvin, beta (37965_at), score: 0.8 PDE1Cphosphodiesterase 1C, calmodulin-dependent 70kDa (207303_at), score: -0.78 PGCprogastricsin (pepsinogen C) (205261_at), score: -0.74 PHF7PHD finger protein 7 (215622_x_at), score: -0.86 PIK3R5phosphoinositide-3-kinase, regulatory subunit 5 (220566_at), score: -0.73 PIP5K1Cphosphatidylinositol-4-phosphate 5-kinase, type I, gamma (212518_at), score: 0.86 PKN1protein kinase N1 (202161_at), score: 0.81 PLAUplasminogen activator, urokinase (211668_s_at), score: 0.75 PNMAL1PNMA-like 1 (218824_at), score: 0.73 POLR2Apolymerase (RNA) II (DNA directed) polypeptide A, 220kDa (202725_at), score: 0.75 POLRMTpolymerase (RNA) mitochondrial (DNA directed) (203782_s_at), score: 0.81 PRKACAprotein kinase, cAMP-dependent, catalytic, alpha (202801_at), score: 0.74 PRKCDprotein kinase C, delta (202545_at), score: 0.84 PRLHprolactin releasing hormone (221443_x_at), score: -0.76 PTPN6protein tyrosine phosphatase, non-receptor type 6 (206687_s_at), score: -0.95 RASGRP3RAS guanyl releasing protein 3 (calcium and DAG-regulated) (205801_s_at), score: 0.72 RICH2Rho-type GTPase-activating protein RICH2 (215232_at), score: -0.74 RNF40ring finger protein 40 (206845_s_at), score: 0.74 RP5-886K2.1neuronal thread protein AD7c-NTP (207953_at), score: -0.77 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: -0.85 RPRD2regulation of nuclear pre-mRNA domain containing 2 (212553_at), score: 0.79 RPS17P5ribosomal protein S17 pseudogene 5 (216348_at), score: -0.73 SBNO2strawberry notch homolog 2 (Drosophila) (204166_at), score: 0.81 SCAMP4secretory carrier membrane protein 4 (213244_at), score: 0.83 SIGLEC1sialic acid binding Ig-like lectin 1, sialoadhesin (44673_at), score: -0.76 SIGLEC6sialic acid binding Ig-like lectin 6 (210796_x_at), score: -0.78 SIN3BSIN3 homolog B, transcription regulator (yeast) (209352_s_at), score: 0.84 SLC1A7solute carrier family 1 (glutamate transporter), member 7 (210923_at), score: -0.8 SLC26A10solute carrier family 26, member 10 (214951_at), score: -0.94 SLC5A5solute carrier family 5 (sodium iodide symporter), member 5 (211123_at), score: -0.83 SLC9A1solute carrier family 9 (sodium/hydrogen exchanger), member 1 (209453_at), score: 0.74 SMG6Smg-6 homolog, nonsense mediated mRNA decay factor (C. elegans) (214940_s_at), score: 0.84 SPATA1spermatogenesis associated 1 (221057_at), score: -0.85 SULT1B1sulfotransferase family, cytosolic, 1B, member 1 (207601_at), score: -0.82 TAOK2TAO kinase 2 (204986_s_at), score: 0.7 TAPBPTAP binding protein (tapasin) (208829_at), score: 0.79 TBX21T-box 21 (220684_at), score: -0.78 TERF2telomeric repeat binding factor 2 (203611_at), score: 0.71 TIAF1TGFB1-induced anti-apoptotic factor 1 (202039_at), score: 0.71 TMCC2transmembrane and coiled-coil domain family 2 (213096_at), score: -0.72 TMSB4Ythymosin beta 4, Y-linked (206769_at), score: -0.87 TULP2tubby like protein 2 (206733_at), score: -0.87 UBTD1ubiquitin domain containing 1 (219172_at), score: 0.82 VEGFBvascular endothelial growth factor B (203683_s_at), score: 0.91 VPS37Bvacuolar protein sorting 37 homolog B (S. cerevisiae) (221704_s_at), score: 0.77 WDR42AWD repeat domain 42A (202249_s_at), score: 0.73 XAB2XPA binding protein 2 (218110_at), score: 0.72 ZFHX3zinc finger homeobox 3 (208033_s_at), score: 0.81 ZNF282zinc finger protein 282 (212892_at), score: 0.73
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3407-raw-cel-1437949557.cel | 1 | 4 | Cockayne | hgu133a | CS | eGFP |
| E-TABM-263-raw-cel-1515486031.cel | 20 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486071.cel | 22 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |