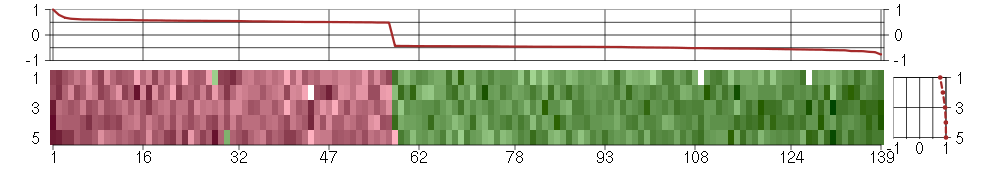

Under-expression is coded with green,
over-expression with red color.


intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
intermediate filament
A cytoskeletal structure that forms a distinct elongated structure, characteristically 10 nm in diameter, that occurs in the cytoplasm of eukaryotic cells. Intermediate filaments form a fibrous system, composed of chemically heterogeneous subunits and involved in mechanically integrating the various components of the cytoplasmic space. Intermediate filaments may be divided into five chemically distinct classes: Type I, acidic keratins; Type II, basic keratins; Type III, including desmin, vimentin and others; Type IV, neurofilaments and related filaments; and Type V, lamins.
extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
extracellular space
That part of a multicellular organism outside the cells proper, usually taken to be outside the plasma membranes, and occupied by fluid.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
cytoskeleton
Any of the various filamentous elements that form the internal framework of cells, and typically remain after treatment of the cells with mild detergent to remove membrane constituents and soluble components of the cytoplasm. The term embraces intermediate filaments, microfilaments, microtubules, the microtrabecular lattice, and other structures characterized by a polymeric filamentous nature and long-range order within the cell. The various elements of the cytoskeleton not only serve in the maintenance of cellular shape but also have roles in other cellular functions, including cellular movement, cell division, endocytosis, and movement of organelles.
extracellular matrix
A structure lying external to one or more cells, which provides structural support for cells or tissues; may be completely external to the cell (as in animals) or be part of the cell (as in plants).
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane. Includes ribosomes, the cytoskeleton and chromosomes.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane and occurring within the cell. Includes ribosomes, the cytoskeleton and chromosomes.
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
cytoskeletal part
Any constituent part of the cytoskeleton, a cellular scaffolding or skeleton that maintains cell shape, enables some cell motion (using structures such as flagella and cilia), and plays important roles in both intra-cellular transport (e.g. the movement of vesicles and organelles) and cellular division. Includes constituent parts of intermediate filaments, microfilaments, microtubules, and the microtrabecular lattice.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
intermediate filament cytoskeleton
Cytoskeletal structure made from intermediate filaments, typically organized in the cytosol as an extended system that stretches from the nuclear envelope to the plasma membrane. Some intermediate filaments run parallel to the cell surface, while others traverse the cytosol; together they form an internal framework that helps support the shape and resilience of the cell.
all
This term is the most general term possible
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
intracellular non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane and occurring within the cell. Includes ribosomes, the cytoskeleton and chromosomes.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
cytoskeletal part
Any constituent part of the cytoskeleton, a cellular scaffolding or skeleton that maintains cell shape, enables some cell motion (using structures such as flagella and cilia), and plays important roles in both intra-cellular transport (e.g. the movement of vesicles and organelles) and cellular division. Includes constituent parts of intermediate filaments, microfilaments, microtubules, and the microtrabecular lattice.
intermediate filament
A cytoskeletal structure that forms a distinct elongated structure, characteristically 10 nm in diameter, that occurs in the cytoplasm of eukaryotic cells. Intermediate filaments form a fibrous system, composed of chemically heterogeneous subunits and involved in mechanically integrating the various components of the cytoplasmic space. Intermediate filaments may be divided into five chemically distinct classes: Type I, acidic keratins; Type II, basic keratins; Type III, including desmin, vimentin and others; Type IV, neurofilaments and related filaments; and Type V, lamins.

molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
catalytic activity
Catalysis of a biochemical reaction at physiological temperatures. In biologically catalyzed reactions, the reactants are known as substrates, and the catalysts are naturally occurring macromolecular substances known as enzymes. Enzymes possess specific binding sites for substrates, and are usually composed wholly or largely of protein, but RNA that has catalytic activity (ribozyme) is often also regarded as enzymatic.
endopeptidase activity
Catalysis of the hydrolysis of internal, alpha-peptide bonds in a polypeptide chain.
peptidase activity
Catalysis of the hydrolysis of a peptide bond. A peptide bond is a covalent bond formed when the carbon atom from the carboxyl group of one amino acid shares electrons with the nitrogen atom from the amino group of a second amino acid.
peptidase activity, acting on L-amino acid peptides
Catalysis of the hydrolysis of peptide bonds formed between L-amino acids.
hydrolase activity
Catalysis of the hydrolysis of various bonds, e.g. C-O, C-N, C-C, phosphoric anhydride bonds, etc. Hydrolase is the systematic name for any enzyme of EC class 3.
all
This term is the most general term possible
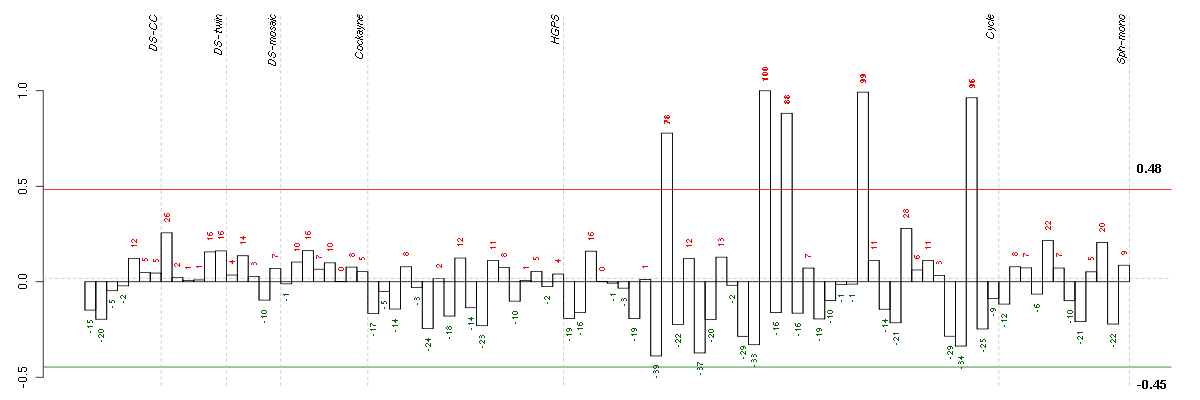
ACANaggrecan (205679_x_at), score: 0.54 ADAMTS6ADAM metallopeptidase with thrombospondin type 1 motif, 6 (220866_at), score: -0.46 ANXA10annexin A10 (210143_at), score: -0.55 APOBEC3Fapolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3F (214995_s_at), score: 0.52 ARMC9armadillo repeat containing 9 (219637_at), score: 0.53 ARNT2aryl-hydrocarbon receptor nuclear translocator 2 (202986_at), score: 0.56 ASPNasporin (219087_at), score: -0.47 ASTN2astrotactin 2 (209693_at), score: 0.57 B3GNTL1UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase-like 1 (213589_s_at), score: 0.55 BAXBCL2-associated X protein (208478_s_at), score: 0.5 BCCIPBRCA2 and CDKN1A interacting protein (218264_at), score: 0.51 C11orf67chromosome 11 open reading frame 67 (221600_s_at), score: 0.53 C17orf39chromosome 17 open reading frame 39 (220058_at), score: 0.58 C19orf54chromosome 19 open reading frame 54 (222052_at), score: 0.51 C1orf9chromosome 1 open reading frame 9 (203429_s_at), score: -0.51 C21orf7chromosome 21 open reading frame 7 (221211_s_at), score: -0.44 C2CD2LC2CD2-like (204757_s_at), score: -0.46 CCL11chemokine (C-C motif) ligand 11 (210133_at), score: -0.49 CD320CD320 molecule (218529_at), score: 0.61 CD55CD55 molecule, decay accelerating factor for complement (Cromer blood group) (201925_s_at), score: -0.54 CDCP1CUB domain containing protein 1 (218451_at), score: -0.5 CDH18cadherin 18, type 2 (206280_at), score: 0.52 CLCF1cardiotrophin-like cytokine factor 1 (219500_at), score: -0.47 CLEC3BC-type lectin domain family 3, member B (205200_at), score: -0.67 CLK1CDC-like kinase 1 (214683_s_at), score: -0.49 CLK4CDC-like kinase 4 (210346_s_at), score: -0.48 CLUclusterin (208791_at), score: -0.56 CNNM2cyclin M2 (206818_s_at), score: 0.49 COL14A1collagen, type XIV, alpha 1 (212865_s_at), score: -0.48 COL5A3collagen, type V, alpha 3 (52255_s_at), score: -0.66 COL7A1collagen, type VII, alpha 1 (217312_s_at), score: -0.47 CORO2Bcoronin, actin binding protein, 2B (209789_at), score: -0.52 CPT1Acarnitine palmitoyltransferase 1A (liver) (203633_at), score: 0.49 CRYL1crystallin, lambda 1 (220753_s_at), score: -0.43 CST6cystatin E/M (206595_at), score: -0.63 CTSKcathepsin K (202450_s_at), score: -0.47 CTSOcathepsin O (203758_at), score: -0.49 CTSScathepsin S (202901_x_at), score: -0.46 CXCL6chemokine (C-X-C motif) ligand 6 (granulocyte chemotactic protein 2) (206336_at), score: -0.46 DCP2DCP2 decapping enzyme homolog (S. cerevisiae) (212919_at), score: -0.45 DIRAS3DIRAS family, GTP-binding RAS-like 3 (215506_s_at), score: -0.6 DNAJB9DnaJ (Hsp40) homolog, subfamily B, member 9 (202843_at), score: -0.46 DPP4dipeptidyl-peptidase 4 (211478_s_at), score: -0.46 DUS4Ldihydrouridine synthase 4-like (S. cerevisiae) (205761_s_at), score: 0.51 DUSP5dual specificity phosphatase 5 (209457_at), score: -0.49 DUSP6dual specificity phosphatase 6 (208891_at), score: -0.56 EDNRBendothelin receptor type B (204271_s_at), score: -0.44 EMCNendomucin (219436_s_at), score: 0.59 EML1echinoderm microtubule associated protein like 1 (204797_s_at), score: -0.57 EVI2Becotropic viral integration site 2B (211742_s_at), score: 0.56 FAM29Afamily with sequence similarity 29, member A (218602_s_at), score: -0.45 FAM86Cfamily with sequence similarity 86, member C (220353_at), score: 0.51 FBXO31F-box protein 31 (219785_s_at), score: 0.52 FGD2FYVE, RhoGEF and PH domain containing 2 (215602_at), score: -0.44 FZD4frizzled homolog 4 (Drosophila) (218665_at), score: 0.6 GAAglucosidase, alpha; acid (202812_at), score: -0.51 GALTgalactose-1-phosphate uridylyltransferase (203179_at), score: 0.54 GKglycerol kinase (207387_s_at), score: -0.5 GPM6Bglycoprotein M6B (209167_at), score: -0.59 GPR177G protein-coupled receptor 177 (221958_s_at), score: -0.53 GPRASP1G protein-coupled receptor associated sorting protein 1 (204793_at), score: 0.68 HGFhepatocyte growth factor (hepapoietin A; scatter factor) (209960_at), score: -0.54 HSPB2heat shock 27kDa protein 2 (205824_at), score: 0.6 HSPBAP1HSPB (heat shock 27kDa) associated protein 1 (219284_at), score: 0.49 IGF2insulin-like growth factor 2 (somatomedin A) (202409_at), score: -0.51 IL13RA2interleukin 13 receptor, alpha 2 (206172_at), score: -0.6 IL33interleukin 33 (209821_at), score: -0.77 KAL1Kallmann syndrome 1 sequence (205206_at), score: -0.52 KCNE4potassium voltage-gated channel, Isk-related family, member 4 (222379_at), score: 0.56 KCTD9potassium channel tetramerisation domain containing 9 (218823_s_at), score: -0.47 KITv-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog (205051_s_at), score: 0.55 KRT17keratin 17 (212236_x_at), score: -0.43 KRT33Akeratin 33A (208483_x_at), score: -0.57 KRT9keratin 9 (208188_at), score: -0.44 LMAN2Llectin, mannose-binding 2-like (221274_s_at), score: 0.57 LOC388152hypothetical LOC388152 (220602_s_at), score: 0.55 LRP8low density lipoprotein receptor-related protein 8, apolipoprotein e receptor (205282_at), score: -0.44 MID1IP1MID1 interacting protein 1 (gastrulation specific G12 homolog (zebrafish)) (218251_at), score: -0.46 MLPHmelanophilin (218211_s_at), score: 0.6 MMP10matrix metallopeptidase 10 (stromelysin 2) (205680_at), score: -0.44 MPImannose phosphate isomerase (202472_at), score: 0.51 MSTP9macrophage stimulating, pseudogene 9 (213382_at), score: -0.52 NAGPAN-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase (205090_s_at), score: 0.56 NEFLneurofilament, light polypeptide (221805_at), score: -0.63 NFATC2IPnuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 interacting protein (217526_at), score: -0.55 NOL8nucleolar protein 8 (218244_at), score: 0.53 NR5A2nuclear receptor subfamily 5, group A, member 2 (208343_s_at), score: -0.44 NTHL1nth endonuclease III-like 1 (E. coli) (209731_at), score: 0.55 NUPL1nucleoporin like 1 (204435_at), score: -0.44 OLFML2Aolfactomedin-like 2A (213075_at), score: -0.53 OPCMLopioid binding protein/cell adhesion molecule-like (214111_at), score: -0.45 PAFAH1B3platelet-activating factor acetylhydrolase, isoform Ib, gamma subunit 29kDa (203228_at), score: 0.5 PENKproenkephalin (213791_at), score: -0.44 PET112LPET112-like (yeast) (204300_at), score: 0.51 PHKA1phosphorylase kinase, alpha 1 (muscle) (205450_at), score: -0.44 PIGLphosphatidylinositol glycan anchor biosynthesis, class L (213889_at), score: 0.52 PKIAprotein kinase (cAMP-dependent, catalytic) inhibitor alpha (204612_at), score: -0.53 PLAURplasminogen activator, urokinase receptor (211924_s_at), score: -0.43 PMAIP1phorbol-12-myristate-13-acetate-induced protein 1 (204285_s_at), score: -0.57 PNRC2proline-rich nuclear receptor coactivator 2 (217779_s_at), score: -0.57 POP1processing of precursor 1, ribonuclease P/MRP subunit (S. cerevisiae) (213449_at), score: 0.5 PRELPproline/arginine-rich end leucine-rich repeat protein (204223_at), score: -0.44 PRKG2protein kinase, cGMP-dependent, type II (207505_at), score: -0.59 PRSS2protease, serine, 2 (trypsin 2) (205402_x_at), score: -0.53 PRSS3protease, serine, 3 (213421_x_at), score: -0.63 PSMB9proteasome (prosome, macropain) subunit, beta type, 9 (large multifunctional peptidase 2) (204279_at), score: 0.59 PTGS1prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) (215813_s_at), score: -0.54 PYCARDPYD and CARD domain containing (221666_s_at), score: 0.5 RAB8BRAB8B, member RAS oncogene family (219210_s_at), score: 0.57 RARRES1retinoic acid receptor responder (tazarotene induced) 1 (221872_at), score: -0.44 RASA2RAS p21 protein activator 2 (206636_at), score: -0.46 RASSF2Ras association (RalGDS/AF-6) domain family member 2 (203185_at), score: -0.48 RBM39RNA binding motif protein 39 (207941_s_at), score: -0.44 RBM4BRNA binding motif protein 4B (209497_s_at), score: 0.56 RP2retinitis pigmentosa 2 (X-linked recessive) (205191_at), score: -0.46 RPL23ribosomal protein L23 (214744_s_at), score: -0.45 SAT1spermidine/spermine N1-acetyltransferase 1 (213988_s_at), score: -0.53 SETMARSET domain and mariner transposase fusion gene (206554_x_at), score: 0.56 SFRS12splicing factor, arginine/serine-rich 12 (212721_at), score: -0.47 SLC16A5solute carrier family 16, member 5 (monocarboxylic acid transporter 6) (206600_s_at), score: 0.62 SLC16A6solute carrier family 16, member 6 (monocarboxylic acid transporter 7) (207038_at), score: -0.46 SLC38A4solute carrier family 38, member 4 (220786_s_at), score: 0.79 SOX9SRY (sex determining region Y)-box 9 (202935_s_at), score: -0.45 SPA17sperm autoantigenic protein 17 (205406_s_at), score: 0.51 SUOXsulfite oxidase (204067_at), score: 0.55 SYNMsynemin, intermediate filament protein (212730_at), score: 0.59 SYT11synaptotagmin XI (209198_s_at), score: 0.61 TFDP2transcription factor Dp-2 (E2F dimerization partner 2) (203588_s_at), score: 0.6 TGFAtransforming growth factor, alpha (205016_at), score: -0.46 TMEM158transmembrane protein 158 (213338_at), score: -0.46 TMSB15Athymosin beta 15a (205347_s_at), score: 0.64 TOXthymocyte selection-associated high mobility group box (204529_s_at), score: 0.5 TPK1thiamin pyrophosphokinase 1 (221218_s_at), score: -0.57 TRIAP1TP53 regulated inhibitor of apoptosis 1 (218403_at), score: 0.53 TRIB2tribbles homolog 2 (Drosophila) (202478_at), score: 0.56 UCP2uncoupling protein 2 (mitochondrial, proton carrier) (208998_at), score: 1 VWA5Avon Willebrand factor A domain containing 5A (210102_at), score: 0.55 XPO4exportin 4 (218479_s_at), score: 0.58 ZNF219zinc finger protein 219 (219314_s_at), score: -0.53
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486051.cel | 21 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486391.cel | 38 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486191.cel | 28 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486011.cel | 19 | 6 | Cycle | hgu133a2 | none | Cycle 1 |