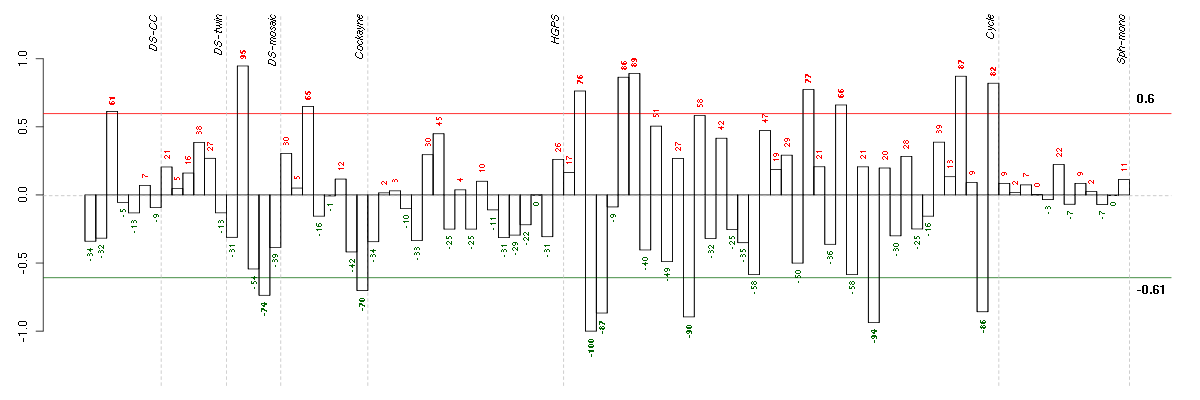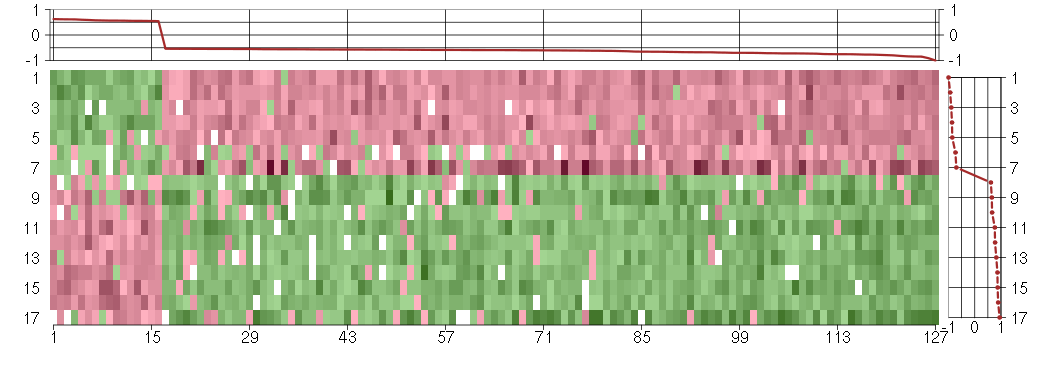

Under-expression is coded with green,
over-expression with red color.



protein binding
Interacting selectively with any protein or protein complex (a complex of two or more proteins that may include other nonprotein molecules).
molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
binding
The selective, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
all
This term is the most general term possible
| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 04010 | 1.039e-02 | 2.519 | 10 | 183 | MAPK signaling pathway |
| 04510 | 4.319e-02 | 2.078 | 8 | 151 | Focal adhesion |
ABCD1ATP-binding cassette, sub-family D (ALD), member 1 (205142_x_at), score: -0.6 ABLIM1actin binding LIM protein 1 (200965_s_at), score: -0.55 ADCK2aarF domain containing kinase 2 (221893_s_at), score: -0.7 ADRBK1adrenergic, beta, receptor kinase 1 (201401_s_at), score: -0.54 ANKFY1ankyrin repeat and FYVE domain containing 1 (219868_s_at), score: -0.57 ARHGDIARho GDP dissociation inhibitor (GDI) alpha (213606_s_at), score: -0.75 ARHGEF11Rho guanine nucleotide exchange factor (GEF) 11 (202914_s_at), score: -0.58 ATF5activating transcription factor 5 (204999_s_at), score: -0.72 ATP6V0A1ATPase, H+ transporting, lysosomal V0 subunit a1 (205095_s_at), score: -0.57 BAT2HLA-B associated transcript 2 (212081_x_at), score: -0.62 BCL2L1BCL2-like 1 (215037_s_at), score: -0.73 BET1Lblocked early in transport 1 homolog (S. cerevisiae)-like (220470_at), score: -0.62 BTBD2BTB (POZ) domain containing 2 (207722_s_at), score: -0.79 C14orf45chromosome 14 open reading frame 45 (220173_at), score: 0.56 C19orf22chromosome 19 open reading frame 22 (221764_at), score: -0.54 C19orf6chromosome 19 open reading frame 6 (213986_s_at), score: -0.65 C19orf61chromosome 19 open reading frame 61 (221335_x_at), score: -0.71 C20orf111chromosome 20 open reading frame 111 (209020_at), score: 0.56 C20orf27chromosome 20 open reading frame 27 (50314_i_at), score: -0.6 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (217849_s_at), score: -0.75 CICcapicua homolog (Drosophila) (212784_at), score: -0.61 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: -0.91 COL6A1collagen, type VI, alpha 1 (212940_at), score: -0.72 DCBLD2discoidin, CUB and LCCL domain containing 2 (213865_at), score: -0.59 DENND3DENN/MADD domain containing 3 (212974_at), score: -0.69 DLSTPdihydrolipoamide S-succinyltransferase pseudogene (E2 component of 2-oxo-glutarate complex) (215210_s_at), score: 0.55 DMWDdystrophia myotonica, WD repeat containing (33768_at), score: -0.71 DRG2developmentally regulated GTP binding protein 2 (203267_s_at), score: -0.66 EHBP1L1EH domain binding protein 1-like 1 (221755_at), score: -0.6 ELK1ELK1, member of ETS oncogene family (210376_x_at), score: -0.68 FBXL18F-box and leucine-rich repeat protein 18 (215068_s_at), score: -0.54 FKRPfukutin related protein (219853_at), score: -0.76 FOXK2forkhead box K2 (203064_s_at), score: -0.67 FRG1FSHD region gene 1 (204145_at), score: 0.54 GPR137G protein-coupled receptor 137 (43934_at), score: -0.56 H1FXH1 histone family, member X (204805_s_at), score: -0.59 HIPK1homeodomain interacting protein kinase 1 (212291_at), score: -0.54 HMGA1high mobility group AT-hook 1 (210457_x_at), score: -0.85 HNRNPUL1heterogeneous nuclear ribonucleoprotein U-like 1 (209675_s_at), score: -0.59 HPS1Hermansky-Pudlak syndrome 1 (217354_s_at), score: -0.61 HSD17B6hydroxysteroid (17-beta) dehydrogenase 6 homolog (mouse) (37512_at), score: 0.61 IDUAiduronidase, alpha-L- (205059_s_at), score: -0.66 ITGA1integrin, alpha 1 (214660_at), score: 0.55 JARID1Cjumonji, AT rich interactive domain 1C (202383_at), score: -0.56 JUNDjun D proto-oncogene (203751_x_at), score: -0.57 JUPjunction plakoglobin (201015_s_at), score: -0.56 KATNB1katanin p80 (WD repeat containing) subunit B 1 (203162_s_at), score: 0.55 KCTD2potassium channel tetramerisation domain containing 2 (212564_at), score: -0.55 KIAA1305KIAA1305 (220911_s_at), score: -0.55 KLHL23kelch-like 23 (Drosophila) (213610_s_at), score: 0.57 LRDDleucine-rich repeats and death domain containing (221640_s_at), score: -0.75 LRP2low density lipoprotein-related protein 2 (205710_at), score: -0.6 LTBP3latent transforming growth factor beta binding protein 3 (219922_s_at), score: -0.56 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: -0.58 MAP2K2mitogen-activated protein kinase kinase 2 (213490_s_at), score: -0.67 MAP3K2mitogen-activated protein kinase kinase kinase 2 (221695_s_at), score: -0.61 MAP7D1MAP7 domain containing 1 (217943_s_at), score: -0.54 MAPKAPK2mitogen-activated protein kinase-activated protein kinase 2 (201461_s_at), score: -0.57 MAZMYC-associated zinc finger protein (purine-binding transcription factor) (207824_s_at), score: -0.68 MED15mediator complex subunit 15 (222175_s_at), score: -0.56 MGAT4Bmannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme B (220189_s_at), score: -0.59 MRPS6mitochondrial ribosomal protein S6 (213167_s_at), score: -0.77 MUC1mucin 1, cell surface associated (207847_s_at), score: -0.57 MYO9Bmyosin IXB (217297_s_at), score: -0.55 NAB2NGFI-A binding protein 2 (EGR1 binding protein 2) (216017_s_at), score: -0.62 NDPNorrie disease (pseudoglioma) (206022_at), score: -0.59 NEFMneurofilament, medium polypeptide (205113_at), score: 0.56 NF1neurofibromin 1 (211094_s_at), score: -0.82 NFATC4nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 (205897_at), score: -0.7 NOTCH2Notch homolog 2 (Drosophila) (210756_s_at), score: -0.61 NUMA1nuclear mitotic apparatus protein 1 (214251_s_at), score: -0.6 NUPL2nucleoporin like 2 (204003_s_at), score: 0.59 OAS32'-5'-oligoadenylate synthetase 3, 100kDa (218400_at), score: -0.69 OGDHoxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) (201282_at), score: -0.55 OLFML2Bolfactomedin-like 2B (213125_at), score: -0.73 ORAI2ORAI calcium release-activated calcium modulator 2 (218812_s_at), score: -0.56 PAK1p21 protein (Cdc42/Rac)-activated kinase 1 (209615_s_at), score: -0.57 PARVBparvin, beta (37965_at), score: -0.55 PCDHGA1protocadherin gamma subfamily A, 1 (209079_x_at), score: -0.59 PCDHGA11protocadherin gamma subfamily A, 11 (211876_x_at), score: -0.8 PCDHGA3protocadherin gamma subfamily A, 3 (216352_x_at), score: -0.85 PCNXpecanex homolog (Drosophila) (213159_at), score: -0.66 PDE5Aphosphodiesterase 5A, cGMP-specific (206757_at), score: -0.55 PEX6peroxisomal biogenesis factor 6 (204545_at), score: -0.57 PHLDA1pleckstrin homology-like domain, family A, member 1 (217997_at), score: -0.59 PLD3phospholipase D family, member 3 (201050_at), score: -0.63 PLEC1plectin 1, intermediate filament binding protein 500kDa (216971_s_at), score: -0.55 POLDIP3polymerase (DNA-directed), delta interacting protein 3 (215357_s_at), score: -0.58 POM121POM121 membrane glycoprotein (rat) (212178_s_at), score: -0.77 PRKCDprotein kinase C, delta (202545_at), score: -0.57 PRMT3protein arginine methyltransferase 3 (213320_at), score: 0.62 PRRX1paired related homeobox 1 (205991_s_at), score: -0.56 PTBP1polypyrimidine tract binding protein 1 (212016_s_at), score: -0.59 PTPROprotein tyrosine phosphatase, receptor type, O (211600_at), score: -0.57 PXNpaxillin (211823_s_at), score: -1 RAB11BRAB11B, member RAS oncogene family (34478_at), score: -0.67 RASA3RAS p21 protein activator 3 (206220_s_at), score: -0.7 RELAv-rel reticuloendotheliosis viral oncogene homolog A (avian) (209878_s_at), score: -0.72 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: 0.62 RPS17P5ribosomal protein S17 pseudogene 5 (216348_at), score: 0.6 RXRBretinoid X receptor, beta (215099_s_at), score: -0.65 SCAMP4secretory carrier membrane protein 4 (213244_at), score: -0.7 SENP3SUMO1/sentrin/SMT3 specific peptidase 3 (215113_s_at), score: -0.68 SF1splicing factor 1 (208313_s_at), score: -0.59 SH3GLB2SH3-domain GRB2-like endophilin B2 (218813_s_at), score: -0.75 SHC1SHC (Src homology 2 domain containing) transforming protein 1 (201469_s_at), score: -0.62 SIRT6sirtuin (silent mating type information regulation 2 homolog) 6 (S. cerevisiae) (219613_s_at), score: -0.78 SLC29A1solute carrier family 29 (nucleoside transporters), member 1 (201801_s_at), score: -0.54 SOCS1suppressor of cytokine signaling 1 (210001_s_at), score: -0.58 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: -0.84 SPG7spastic paraplegia 7 (pure and complicated autosomal recessive) (202104_s_at), score: -0.57 SPPL2Bsignal peptide peptidase-like 2B (215833_s_at), score: -0.6 TAPBPTAP binding protein (tapasin) (208829_at), score: -0.59 TGFBR2transforming growth factor, beta receptor II (70/80kDa) (207334_s_at), score: -0.72 TOM1L2target of myb1-like 2 (chicken) (214840_at), score: -0.58 TRIM3tripartite motif-containing 3 (204911_s_at), score: -0.57 TSKUtsukushin (218245_at), score: -0.58 TXLNAtaxilin alpha (212300_at), score: -0.64 UBTD1ubiquitin domain containing 1 (219172_at), score: -0.72 ULK1unc-51-like kinase 1 (C. elegans) (209333_at), score: -0.59 VEGFBvascular endothelial growth factor B (203683_s_at), score: -0.75 WDR42AWD repeat domain 42A (202249_s_at), score: -0.65 WDR6WD repeat domain 6 (217734_s_at), score: -0.59 YAF2YY1 associated factor 2 (206238_s_at), score: 0.56 YWHAEtyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide (210317_s_at), score: -0.57 ZNF281zinc finger protein 281 (218401_s_at), score: 0.58
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486411.cel | 39 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 47B.CEL | 4 | 3 | DS-mosaic | hgu133plus2 | Down mosaic | DS-mosaic 4 |
| E-GEOD-3407-raw-cel-1437949938.cel | 8 | 4 | Cockayne | hgu133a | none | CSB |
| ctrl c 08-03.CEL | 3 | 1 | DS-CC | hgu133a | none | DS-CC 3 |
| E-GEOD-3407-raw-cel-1437949655.cel | 3 | 4 | Cockayne | hgu133a | none | CSB |
| E-TABM-263-raw-cel-1515486151.cel | 26 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 46B.CEL | 2 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 2 |