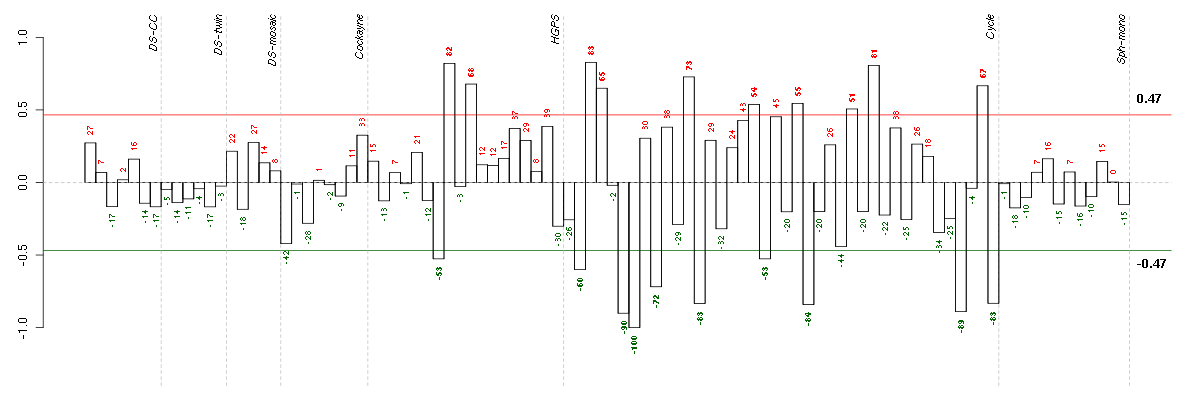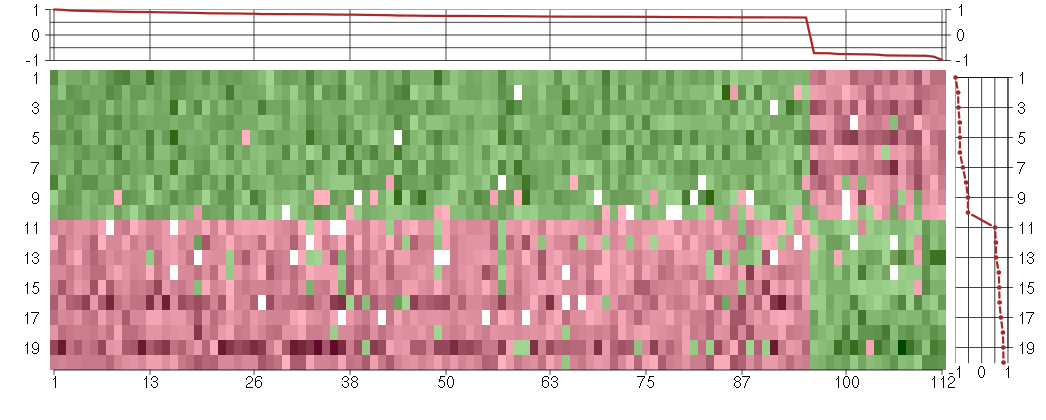

Under-expression is coded with green,
over-expression with red color.

biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
positive regulation of kinase activity
Any process that activates or increases the frequency, rate or extent of kinase activity, the catalysis of the transfer of a phosphate group, usually from ATP, to a substrate molecule.
positive regulation of catalytic activity
Any process that activates or increases the activity of an enzyme.
regulation of MAP kinase activity
Any process that modulates the frequency, rate or extent of MAP kinase activity.
positive regulation of MAP kinase activity
Any process that activates or increases the frequency, rate or extent of MAP kinase activity.
regulation of JUN kinase activity
Any process that modulates the frequency, rate or extent of JUN kinase activity.
positive regulation of JUN kinase activity
Any process that activates or increases the frequency, rate or extent of JUN kinase activity.
regulation of kinase activity
Any process that modulates the frequency, rate or extent of kinase activity, the catalysis of the transfer of a phosphate group, usually from ATP, to a substrate molecule.
regulation of protein kinase activity
Any process that modulates the frequency, rate or extent of protein kinase activity.
positive regulation of protein kinase activity
Any process that activates or increases the frequency, rate or extent of protein kinase activity.
regulation of catalytic activity
Any process that modulates the activity of an enzyme.
regulation of transferase activity
Any process that modulates the frequency, rate or extent of transferase activity, the catalysis of the transfer of a group, e.g. a methyl group, glycosyl group, acyl group, phosphorus-containing, or other groups, from one compound (generally regarded as the donor) to another compound (generally regarded as the acceptor). Transferase is the systematic name for any enzyme of EC class 2.
positive regulation of transferase activity
Any process that activates or increases the frequency, rate or extent of transferase activity, the catalysis of the transfer of a group, e.g. a methyl group, glycosyl group, acyl group, phosphorus-containing, or other groups, from a donor compound to an acceptor.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
regulation of molecular function
Any process that modulates the frequency, rate or extent of molecular functions. Molecular functions are elemental biological activities occurring at the molecular level, such as catalysis or binding.
all
This term is the most general term possible
positive regulation of transferase activity
Any process that activates or increases the frequency, rate or extent of transferase activity, the catalysis of the transfer of a group, e.g. a methyl group, glycosyl group, acyl group, phosphorus-containing, or other groups, from a donor compound to an acceptor.
positive regulation of kinase activity
Any process that activates or increases the frequency, rate or extent of kinase activity, the catalysis of the transfer of a phosphate group, usually from ATP, to a substrate molecule.
positive regulation of protein kinase activity
Any process that activates or increases the frequency, rate or extent of protein kinase activity.
positive regulation of MAP kinase activity
Any process that activates or increases the frequency, rate or extent of MAP kinase activity.
positive regulation of JUN kinase activity
Any process that activates or increases the frequency, rate or extent of JUN kinase activity.


| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 05050 | 2.332e-02 | 0.1458 | 3 | 14 | Dentatorubropallidoluysian atrophy (DRPLA) |
ABCD1ATP-binding cassette, sub-family D (ALD), member 1 (205142_x_at), score: 0.82 ADCK2aarF domain containing kinase 2 (221893_s_at), score: 0.89 ARSAarylsulfatase A (204443_at), score: 0.72 ATF5activating transcription factor 5 (204999_s_at), score: 0.71 ATN1atrophin 1 (40489_at), score: 0.82 B3GAT3beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) (203452_at), score: 0.82 BACE2beta-site APP-cleaving enzyme 2 (217867_x_at), score: 0.72 BAIAP2BAI1-associated protein 2 (205294_at), score: 0.69 BAT2HLA-B associated transcript 2 (212081_x_at), score: 0.82 BTBD2BTB (POZ) domain containing 2 (207722_s_at), score: 0.9 C16orf57chromosome 16 open reading frame 57 (218060_s_at), score: 0.72 C19orf61chromosome 19 open reading frame 61 (221335_x_at), score: 0.72 CABIN1calcineurin binding protein 1 (37652_at), score: 0.7 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (217849_s_at), score: 0.96 CDC42EP1CDC42 effector protein (Rho GTPase binding) 1 (204693_at), score: 0.7 CDC42EP4CDC42 effector protein (Rho GTPase binding) 4 (218062_x_at), score: 0.7 CICcapicua homolog (Drosophila) (212784_at), score: 0.91 CITcitron (rho-interacting, serine/threonine kinase 21) (212801_at), score: 0.8 CIZ1CDKN1A interacting zinc finger protein 1 (205516_x_at), score: 0.84 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: 0.95 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: 0.88 CScitrate synthase (208660_at), score: 0.7 DMWDdystrophia myotonica, WD repeat containing (33768_at), score: 0.84 DNAJB9DnaJ (Hsp40) homolog, subfamily B, member 9 (202843_at), score: -0.71 EHBP1L1EH domain binding protein 1-like 1 (221755_at), score: 0.72 FAM18Bfamily with sequence similarity 18, member B (218446_s_at), score: -0.77 FLJ12529pre-mRNA cleavage factor I, 59 kDa subunit (217866_at), score: 0.9 FOSL1FOS-like antigen 1 (204420_at), score: 0.75 FOXK2forkhead box K2 (203064_s_at), score: 0.91 GRINAglutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) (212090_at), score: 0.75 H1FXH1 histone family, member X (204805_s_at), score: 0.73 HMGA1high mobility group AT-hook 1 (210457_x_at), score: 0.85 HNRNPH2heterogeneous nuclear ribonucleoprotein H2 (H') (201132_at), score: -0.81 HNRNPUL1heterogeneous nuclear ribonucleoprotein U-like 1 (209675_s_at), score: 0.75 HSD17B6hydroxysteroid (17-beta) dehydrogenase 6 homolog (mouse) (37512_at), score: -0.76 HSPA12Aheat shock 70kDa protein 12A (214434_at), score: 0.75 IDUAiduronidase, alpha-L- (205059_s_at), score: 0.84 KAL1Kallmann syndrome 1 sequence (205206_at), score: -0.8 LEPREL2leprecan-like 2 (204854_at), score: 0.74 LMF2lipase maturation factor 2 (212682_s_at), score: 0.75 LOC100132540similar to LOC339047 protein (214870_x_at), score: 0.74 LOC339047hypothetical protein LOC339047 (221501_x_at), score: 0.8 LOC391132similar to hCG2041276 (216177_at), score: -0.75 LOC90379hypothetical protein BC002926 (221849_s_at), score: 0.7 LRDDleucine-rich repeats and death domain containing (221640_s_at), score: 0.78 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: 0.99 MAP3K6mitogen-activated protein kinase kinase kinase 6 (219278_at), score: 0.82 MAP7D1MAP7 domain containing 1 (217943_s_at), score: 0.83 MAPKAPK3mitogen-activated protein kinase-activated protein kinase 3 (202788_at), score: 0.72 MAST2microtubule associated serine/threonine kinase 2 (211593_s_at), score: 0.81 MED15mediator complex subunit 15 (222175_s_at), score: 0.75 MFAP3Lmicrofibrillar-associated protein 3-like (205442_at), score: -0.75 MINK1misshapen-like kinase 1 (zebrafish) (214246_x_at), score: 0.75 MLL4myeloid/lymphoid or mixed-lineage leukemia 4 (203419_at), score: 0.69 MUL1mitochondrial E3 ubiquitin ligase 1 (218246_at), score: 0.72 NBL1neuroblastoma, suppression of tumorigenicity 1 (37005_at), score: 0.89 NDPNorrie disease (pseudoglioma) (206022_at), score: 0.69 NEFMneurofilament, medium polypeptide (205113_at), score: -0.81 NFATC4nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 (205897_at), score: 0.85 NPIPnuclear pore complex interacting protein (204538_x_at), score: 0.81 OLFML2Bolfactomedin-like 2B (213125_at), score: 0.87 PAK1p21 protein (Cdc42/Rac)-activated kinase 1 (209615_s_at), score: 0.69 PARVBparvin, beta (37965_at), score: 0.87 PCDHG@protocadherin gamma cluster (215836_s_at), score: 0.75 PCDHGA1protocadherin gamma subfamily A, 1 (209079_x_at), score: 0.85 PEX6peroxisomal biogenesis factor 6 (204545_at), score: 0.69 PIP5K1Cphosphatidylinositol-4-phosphate 5-kinase, type I, gamma (212518_at), score: 0.9 PKN1protein kinase N1 (202161_at), score: 0.71 PLCD1phospholipase C, delta 1 (205125_at), score: 0.72 PNPLA2patatin-like phospholipase domain containing 2 (212705_x_at), score: 0.72 POLR2Apolymerase (RNA) II (DNA directed) polypeptide A, 220kDa (202725_at), score: 0.78 POLSpolymerase (DNA directed) sigma (202466_at), score: 0.71 POM121POM121 membrane glycoprotein (rat) (212178_s_at), score: 0.88 POMZP3POM (POM121 homolog, rat) and ZP3 fusion (204148_s_at), score: 0.73 PRKCDprotein kinase C, delta (202545_at), score: 0.69 PRMT3protein arginine methyltransferase 3 (213320_at), score: -0.8 PTPROprotein tyrosine phosphatase, receptor type, O (211600_at), score: 0.74 PXNpaxillin (211823_s_at), score: 0.79 RAB1ARAB1A, member RAS oncogene family (213440_at), score: -0.71 RAD54L2RAD54-like 2 (S. cerevisiae) (213205_s_at), score: 0.71 RBM15BRNA binding motif protein 15B (202689_at), score: 0.72 REREarginine-glutamic acid dipeptide (RE) repeats (200940_s_at), score: 0.72 RNF220ring finger protein 220 (219988_s_at), score: 0.93 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: -0.98 RPRD2regulation of nuclear pre-mRNA domain containing 2 (212553_at), score: 0.74 RPS17P5ribosomal protein S17 pseudogene 5 (216348_at), score: -0.85 SBNO2strawberry notch homolog 2 (Drosophila) (204166_at), score: 0.69 SCAMP4secretory carrier membrane protein 4 (213244_at), score: 1 SEMA3Fsema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F (35666_at), score: -0.72 SF1splicing factor 1 (208313_s_at), score: 0.95 SIRT6sirtuin (silent mating type information regulation 2 homolog) 6 (S. cerevisiae) (219613_s_at), score: 0.76 SLC43A3solute carrier family 43, member 3 (213113_s_at), score: 0.69 SMG6Smg-6 homolog, nonsense mediated mRNA decay factor (C. elegans) (214940_s_at), score: 0.71 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: 0.93 STRN4striatin, calmodulin binding protein 4 (217903_at), score: 0.8 SULT1B1sulfotransferase family, cytosolic, 1B, member 1 (207601_at), score: -0.76 TAPBPTAP binding protein (tapasin) (208829_at), score: 0.81 TERF2telomeric repeat binding factor 2 (203611_at), score: 0.69 TICAM2toll-like receptor adaptor molecule 2 (214658_at), score: -0.81 TSKUtsukushin (218245_at), score: 0.7 TXLNAtaxilin alpha (212300_at), score: 0.79 UBTD1ubiquitin domain containing 1 (219172_at), score: 0.8 ULK1unc-51-like kinase 1 (C. elegans) (209333_at), score: 0.73 VEGFBvascular endothelial growth factor B (203683_s_at), score: 0.93 VPS37Bvacuolar protein sorting 37 homolog B (S. cerevisiae) (221704_s_at), score: 0.82 WDR42AWD repeat domain 42A (202249_s_at), score: 0.7 WDR6WD repeat domain 6 (217734_s_at), score: 0.76 XAB2XPA binding protein 2 (218110_at), score: 0.72 YTHDC2YTH domain containing 2 (213077_at), score: -0.74 ZFP36L1zinc finger protein 36, C3H type-like 1 (211965_at), score: 0.76 ZNF536zinc finger protein 536 (206403_at), score: 0.69
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690272.cel | 7 | 5 | HGPS | hgu133a | HGPS | AG11498 |
| E-TABM-263-raw-cel-1515486011.cel | 19 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486171.cel | 27 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485991.cel | 18 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486071.cel | 22 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486411.cel | 39 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690344.cel | 10 | 5 | HGPS | hgu133a | none | GM00038C |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690304.cel | 8 | 5 | HGPS | hgu133a | none | GMO8398C |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |