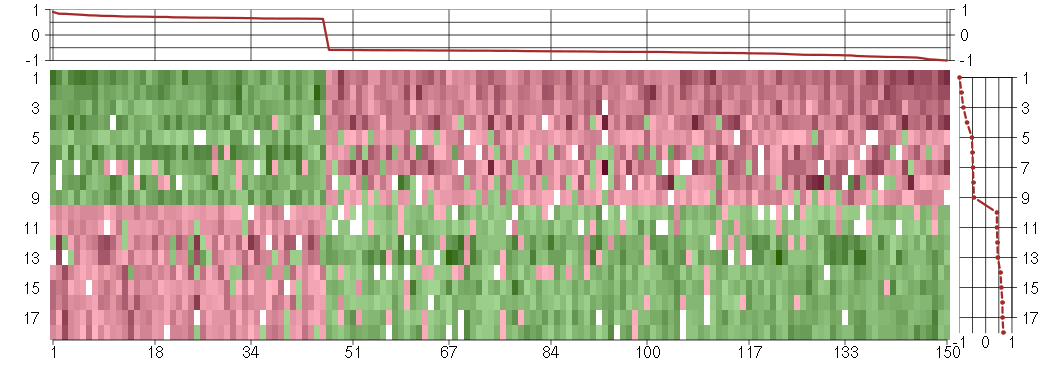

Under-expression is coded with green,
over-expression with red color.

multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
tissue development
The process whose specific outcome is the progression of a tissue over time, from its formation to the mature structure.
ectoderm development
The process whose specific outcome is the progression of the ectoderm over time, from its formation to the mature structure. In animal embryos, the ectoderm is the outer germ layer of the embryo, formed during gastrulation.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cell proliferation
The multiplication or reproduction of cells, resulting in the expansion of a cell population.
epidermis development
The process whose specific outcome is the progression of the epidermis over time, from its formation to the mature structure. The epidermis is the outer epithelial layer of a plant or animal, it may be a single layer that produces an extracellular material (e.g. the cuticle of arthropods) or a complex stratified squamous epithelium, as in the case of many vertebrate species.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
developmental process
A biological process whose specific outcome is the progression of an integrated living unit: an anatomical structure (which may be a subcellular structure, cell, tissue, or organ), or organism over time from an initial condition to a later condition.
regulation of cell proliferation
Any process that modulates the frequency, rate or extent of cell proliferation.
organ development
Development of a tissue or tissues that work together to perform a specific function or functions. Development pertains to the process whose specific outcome is the progression of a structure over time, from its formation to the mature structure. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
anatomical structure development
The biological process whose specific outcome is the progression of an anatomical structure from an initial condition to its mature state. This process begins with the formation of the structure and ends with the mature structure, whatever form that may be including its natural destruction. An anatomical structure is any biological entity that occupies space and is distinguished from its surroundings. Anatomical structures can be macroscopic such as a carpel, or microscopic such as an acrosome.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
all
This term is the most general term possible
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of cell proliferation
Any process that modulates the frequency, rate or extent of cell proliferation.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
organ development
Development of a tissue or tissues that work together to perform a specific function or functions. Development pertains to the process whose specific outcome is the progression of a structure over time, from its formation to the mature structure. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
tissue development
The process whose specific outcome is the progression of a tissue over time, from its formation to the mature structure.
epidermis development
The process whose specific outcome is the progression of the epidermis over time, from its formation to the mature structure. The epidermis is the outer epithelial layer of a plant or animal, it may be a single layer that produces an extracellular material (e.g. the cuticle of arthropods) or a complex stratified squamous epithelium, as in the case of many vertebrate species.

plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
extracellular space
That part of a multicellular organism outside the cells proper, usually taken to be outside the plasma membranes, and occupied by fluid.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.

purinergic nucleotide receptor activity
Combining with a purine nucleotide to initiate a change in cell activity.
molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
signal transducer activity
Mediates the transfer of a signal from the outside to the inside of a cell by means other than the introduction of the signal molecule itself into the cell.
receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity.
transmembrane receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity, and spanning to the membrane of either the cell or an organelle.
nucleotide receptor activity
Combining with a nucleotide to initiate a change in cell activity. A nucleotide is a compound that consists of a nucleoside esterified with a phosphate molecule.
molecular transducer activity
The molecular function that accepts an input of one form and creates an output of a different form.
all
This term is the most general term possible
| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 04080 | 7.794e-03 | 1.48 | 8 | 78 | Neuroactive ligand-receptor interaction |
ABHD5abhydrolase domain containing 5 (218739_at), score: -0.66 ACSL4acyl-CoA synthetase long-chain family member 4 (202422_s_at), score: -0.66 ACTR3BARP3 actin-related protein 3 homolog B (yeast) (218868_at), score: -0.7 ADAM19ADAM metallopeptidase domain 19 (meltrin beta) (209765_at), score: -0.66 ADORA2Badenosine A2b receptor (205891_at), score: 0.8 AGBL5ATP/GTP binding protein-like 5 (218480_at), score: 0.64 AHI1Abelson helper integration site 1 (221569_at), score: -0.87 AIM1absent in melanoma 1 (212543_at), score: 0.77 AJAP1adherens junctions associated protein 1 (206460_at), score: -0.69 ALDH3A2aldehyde dehydrogenase 3 family, member A2 (202053_s_at), score: 0.69 ANXA10annexin A10 (210143_at), score: -0.83 ARL4CADP-ribosylation factor-like 4C (202207_at), score: 0.65 ARMC9armadillo repeat containing 9 (219637_at), score: 0.64 ARNT2aryl-hydrocarbon receptor nuclear translocator 2 (202986_at), score: 0.66 ATP6V0E1ATPase, H+ transporting, lysosomal 9kDa, V0 subunit e1 (214149_s_at), score: 0.65 BMP6bone morphogenetic protein 6 (206176_at), score: -1 C14orf45chromosome 14 open reading frame 45 (220173_at), score: -0.71 C2CD2C2 calcium-dependent domain containing 2 (212875_s_at), score: -0.71 C2CD2LC2CD2-like (204757_s_at), score: -0.64 C5orf23chromosome 5 open reading frame 23 (219054_at), score: -0.65 CALB2calbindin 2 (205428_s_at), score: -0.78 CCDC28Acoiled-coil domain containing 28A (209479_at), score: 0.75 CD55CD55 molecule, decay accelerating factor for complement (Cromer blood group) (201925_s_at), score: -0.76 CDC2L6cell division cycle 2-like 6 (CDK8-like) (212899_at), score: 0.71 CDCP1CUB domain containing protein 1 (218451_at), score: -0.66 CEP68centrosomal protein 68kDa (212677_s_at), score: 0.71 CEP70centrosomal protein 70kDa (219036_at), score: 0.67 COL5A3collagen, type V, alpha 3 (52255_s_at), score: -0.61 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: 0.67 CSGALNACT2chondroitin sulfate N-acetylgalactosaminyltransferase 2 (222235_s_at), score: -0.66 CST6cystatin E/M (206595_at), score: -0.78 CXCL6chemokine (C-X-C motif) ligand 6 (granulocyte chemotactic protein 2) (206336_at), score: -0.77 DCP2DCP2 decapping enzyme homolog (S. cerevisiae) (212919_at), score: -0.6 DENND5BDENN/MADD domain containing 5B (215058_at), score: -0.66 DEPDC6DEP domain containing 6 (218858_at), score: 0.75 DIRAS3DIRAS family, GTP-binding RAS-like 3 (215506_s_at), score: -0.82 DIS3DIS3 mitotic control homolog (S. cerevisiae) (218362_s_at), score: -0.65 DNAJB9DnaJ (Hsp40) homolog, subfamily B, member 9 (202843_at), score: -0.91 DUSP4dual specificity phosphatase 4 (204014_at), score: -0.72 DZIP3DAZ interacting protein 3, zinc finger (213186_at), score: 0.73 EDEM1ER degradation enhancer, mannosidase alpha-like 1 (203279_at), score: -0.62 EIF4Eeukaryotic translation initiation factor 4E (201436_at), score: -0.64 ENTPD7ectonucleoside triphosphate diphosphohydrolase 7 (220153_at), score: -0.62 ESM1endothelial cell-specific molecule 1 (208394_x_at), score: -0.88 ESRRAestrogen-related receptor alpha (1487_at), score: -0.7 FAM155Afamily with sequence similarity 155, member A (214825_at), score: -0.59 FBXO31F-box protein 31 (219785_s_at), score: 0.68 FERMT1fermitin family homolog 1 (Drosophila) (218796_at), score: -0.84 FGF1fibroblast growth factor 1 (acidic) (205117_at), score: -0.59 FGF7fibroblast growth factor 7 (keratinocyte growth factor) (205782_at), score: -0.7 FGF9fibroblast growth factor 9 (glia-activating factor) (206404_at), score: 0.68 FICDFIC domain containing (219910_at), score: -0.68 FNBP1formin binding protein 1 (212288_at), score: 0.71 FSTL3follistatin-like 3 (secreted glycoprotein) (203592_s_at), score: -0.6 GALgalanin prepropeptide (214240_at), score: -0.61 GFOD1glucose-fructose oxidoreductase domain containing 1 (219821_s_at), score: -0.66 GKglycerol kinase (207387_s_at), score: -0.96 GK3Pglycerol kinase 3 pseudogene (215966_x_at), score: -0.99 GMCL1germ cell-less homolog 1 (Drosophila) (218458_at), score: -0.62 GPR177G protein-coupled receptor 177 (221958_s_at), score: -0.73 GPR183G protein-coupled receptor 183 (205419_at), score: -0.66 GPSM2G-protein signaling modulator 2 (AGS3-like, C. elegans) (221922_at), score: 0.64 GRB14growth factor receptor-bound protein 14 (206204_at), score: -0.79 GTF2A1Lgeneral transcription factor IIA, 1-like (213413_at), score: 0.83 HDGFhepatoma-derived growth factor (high-mobility group protein 1-like) (200896_x_at), score: 0.68 HOMER2homer homolog 2 (Drosophila) (217080_s_at), score: -0.62 HTR2A5-hydroxytryptamine (serotonin) receptor 2A (207135_at), score: -0.87 IL11interleukin 11 (206924_at), score: -0.74 IL13RA2interleukin 13 receptor, alpha 2 (206172_at), score: -0.79 IL1RL1interleukin 1 receptor-like 1 (207526_s_at), score: -0.61 IL33interleukin 33 (209821_at), score: -0.97 KAL1Kallmann syndrome 1 sequence (205206_at), score: -0.86 KCNG1potassium voltage-gated channel, subfamily G, member 1 (214595_at), score: -0.69 KIAA1462KIAA1462 (213316_at), score: 0.68 KLHL21kelch-like 21 (Drosophila) (203068_at), score: -0.61 LRP8low density lipoprotein receptor-related protein 8, apolipoprotein e receptor (205282_at), score: -0.72 MBOAT2membrane bound O-acyltransferase domain containing 2 (213288_at), score: -0.61 MFAP3Lmicrofibrillar-associated protein 3-like (205442_at), score: -0.6 MMP10matrix metallopeptidase 10 (stromelysin 2) (205680_at), score: -0.85 MMP16matrix metallopeptidase 16 (membrane-inserted) (207012_at), score: -0.65 MREGmelanoregulin (219648_at), score: -0.66 MTAPmethylthioadenosine phosphorylase (211363_s_at), score: -0.6 MTMR9myotubularin related protein 9 (204837_at), score: -0.59 NCALDneurocalcin delta (211685_s_at), score: -0.88 NDUFA2NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2, 8kDa (213550_s_at), score: 0.72 NEFLneurofilament, light polypeptide (221805_at), score: -0.66 NEFMneurofilament, medium polypeptide (205113_at), score: -0.8 NOX4NADPH oxidase 4 (219773_at), score: -0.7 NPnucleoside phosphorylase (201695_s_at), score: -0.61 NPIPnuclear pore complex interacting protein (204538_x_at), score: 0.64 NPTX1neuronal pentraxin I (204684_at), score: -0.74 NR5A2nuclear receptor subfamily 5, group A, member 2 (208343_s_at), score: -0.67 NTMneurotrimin (222020_s_at), score: -0.66 NUPL1nucleoporin like 1 (204435_at), score: -0.6 OPCMLopioid binding protein/cell adhesion molecule-like (214111_at), score: -0.7 P2RX5purinergic receptor P2X, ligand-gated ion channel, 5 (210448_s_at), score: -0.73 P2RY5purinergic receptor P2Y, G-protein coupled, 5 (218589_at), score: 0.66 PCDH9protocadherin 9 (219737_s_at), score: -0.86 PCTK2PCTAIRE protein kinase 2 (221918_at), score: -0.59 PDCD1LG2programmed cell death 1 ligand 2 (220049_s_at), score: -0.59 PDE5Aphosphodiesterase 5A, cGMP-specific (206757_at), score: 0.65 PECRperoxisomal trans-2-enoyl-CoA reductase (221142_s_at), score: 0.64 PET112LPET112-like (yeast) (204300_at), score: 0.64 PIGHphosphatidylinositol glycan anchor biosynthesis, class H (209625_at), score: -0.63 PIGLphosphatidylinositol glycan anchor biosynthesis, class L (213889_at), score: 0.72 PIONpigeon homolog (Drosophila) (222150_s_at), score: -0.6 PLAURplasminogen activator, urokinase receptor (211924_s_at), score: -0.6 PPP2R1Bprotein phosphatase 2 (formerly 2A), regulatory subunit A, beta isoform (202884_s_at), score: -0.58 PRSS2protease, serine, 2 (trypsin 2) (205402_x_at), score: -0.62 PRSS3protease, serine, 3 (213421_x_at), score: -0.68 PTGS1prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) (215813_s_at), score: -0.67 PTGS2prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) (204748_at), score: -0.72 PTHLHparathyroid hormone-like hormone (211756_at), score: -0.85 PTP4A1protein tyrosine phosphatase type IVA, member 1 (200730_s_at), score: -0.61 PTPRBprotein tyrosine phosphatase, receptor type, B (205846_at), score: -0.76 RFX5regulatory factor X, 5 (influences HLA class II expression) (202963_at), score: 0.82 RNF44ring finger protein 44 (203286_at), score: 0.77 RPL23ribosomal protein L23 (214744_s_at), score: -0.64 S1PR1sphingosine-1-phosphate receptor 1 (204642_at), score: -0.6 SERPINB2serpin peptidase inhibitor, clade B (ovalbumin), member 2 (204614_at), score: -0.6 SLC38A4solute carrier family 38, member 4 (220786_s_at), score: 0.73 SLC4A4solute carrier family 4, sodium bicarbonate cotransporter, member 4 (203908_at), score: -0.62 SLC4A7solute carrier family 4, sodium bicarbonate cotransporter, member 7 (209884_s_at), score: -0.78 SLMO2slowmo homolog 2 (Drosophila) (217851_s_at), score: -0.64 SMAD3SMAD family member 3 (218284_at), score: 0.79 SPA17sperm autoantigenic protein 17 (205406_s_at), score: 0.69 SSTR1somatostatin receptor 1 (208482_at), score: -0.68 ST5suppression of tumorigenicity 5 (202440_s_at), score: 0.67 STARD5StAR-related lipid transfer (START) domain containing 5 (213820_s_at), score: 0.69 TENC1tensin like C1 domain containing phosphatase (tensin 2) (212494_at), score: 0.64 TFDP2transcription factor Dp-2 (E2F dimerization partner 2) (203588_s_at), score: 0.9 TGDSTDP-glucose 4,6-dehydratase (208249_s_at), score: -0.64 TGFAtransforming growth factor, alpha (205016_at), score: -0.78 THYN1thymocyte nuclear protein 1 (218491_s_at), score: 0.72 TMEM2transmembrane protein 2 (218113_at), score: -0.8 TMSB15Athymosin beta 15a (205347_s_at), score: 0.68 TMSB15Bthymosin beta 15B (214051_at), score: 0.63 TP53tumor protein p53 (201746_at), score: 0.75 TSPAN13tetraspanin 13 (217979_at), score: -0.63 UCP2uncoupling protein 2 (mitochondrial, proton carrier) (208998_at), score: 0.83 UFM1ubiquitin-fold modifier 1 (218050_at), score: -0.63 UHRF1BP1LUHRF1 binding protein 1-like (213118_at), score: -0.61 ULBP2UL16 binding protein 2 (221291_at), score: -0.61 URB1URB1 ribosome biogenesis 1 homolog (S. cerevisiae) (212996_s_at), score: -0.59 VWA5Avon Willebrand factor A domain containing 5A (210102_at), score: 0.72 WDR6WD repeat domain 6 (217734_s_at), score: 0.67 XYLT1xylosyltransferase I (213725_x_at), score: -0.64 YDD19YDD19 protein (37079_at), score: -0.61 ZDHHC7zinc finger, DHHC-type containing 7 (218606_at), score: 0.64 ZFYVE16zinc finger, FYVE domain containing 16 (203651_at), score: -0.64
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
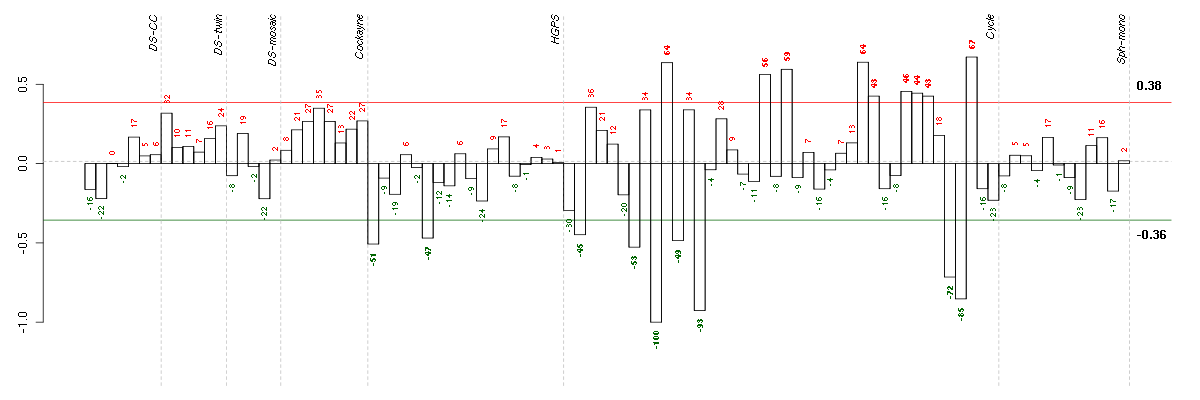
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486351.cel | 36 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690199.cel | 1 | 5 | HGPS | hgu133a | none | GM0316B |
| E-TABM-263-raw-cel-1515485851.cel | 11 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690256.cel | 6 | 5 | HGPS | hgu133a | none | GMO8398C |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486311.cel | 34 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486291.cel | 33 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486271.cel | 32 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486011.cel | 19 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486051.cel | 21 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486191.cel | 28 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486391.cel | 38 | 6 | Cycle | hgu133a2 | none | Cycle 1 |