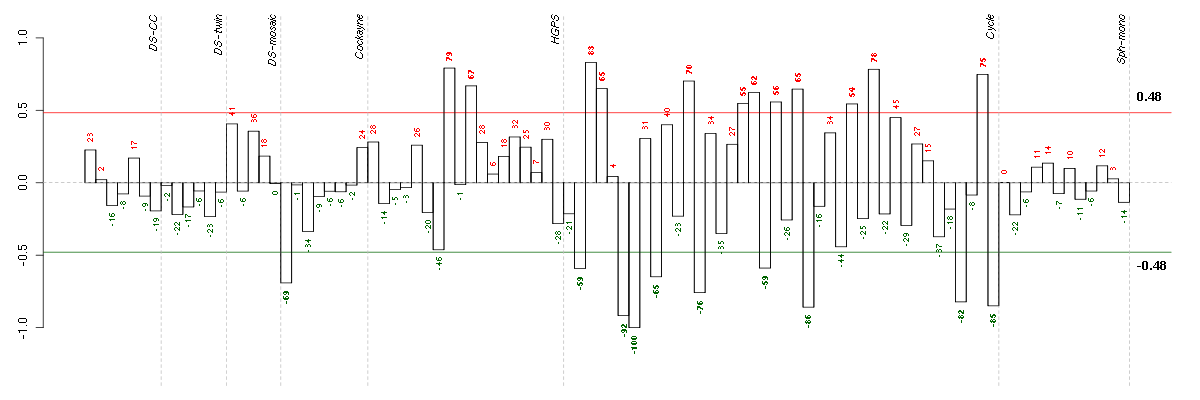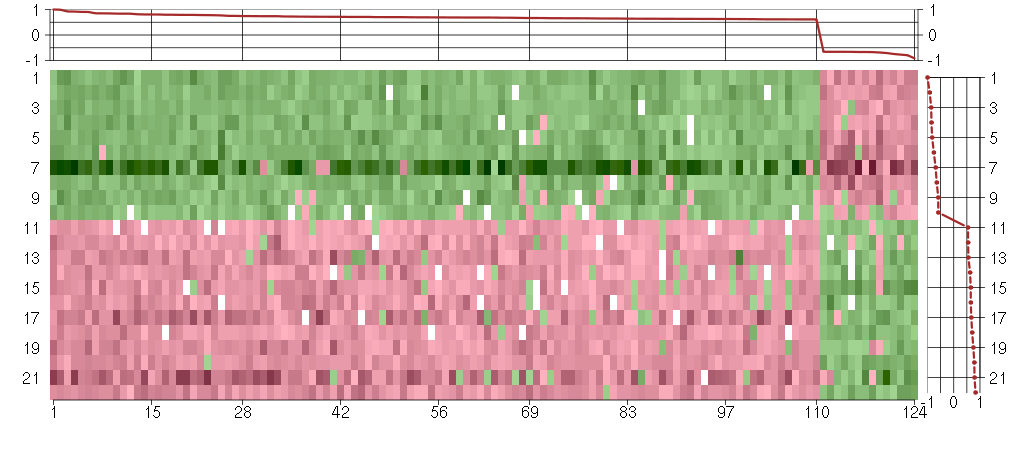

Under-expression is coded with green,
over-expression with red color.

metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
positive regulation of metabolic process
Any process that activates or increases the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism.
gene expression
The process by which a gene's coding sequence is converted into a mature gene product or products (proteins or RNA). This includes the production of an RNA transcript as well as any processing to produce a mature RNA product or an mRNA (for protein-coding genes) and the translation of that mRNA into protein. Some protein processing events may be included when they are required to form an active form of a product from an inactive precursor form.
regulation of gene expression
Any process that modulates the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product or products (proteins or RNA). This includes the production of an RNA transcript as well as any processing to produce a mature RNA product or an mRNA (for protein-coding genes) and the translation of that mRNA into protein. Some protein processing events may be included when they are required to form an active form of a product from an inactive precursor form.
positive regulation of macromolecule metabolic process
Any process that increases the frequency, rate or extent of the chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
positive regulation of gene expression
Any process that increases the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product or products (proteins or RNA). This includes the production of an RNA transcript as well as any processing to produce a mature RNA product or an mRNA (for protein-coding genes) and the translation of that mRNA into protein. Some protein processing events may be included when they are required to form an active form of a product from an inactive precursor form.
regulation of metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism.
regulation of gene expression, epigenetic
Any process that modulates the frequency, rate or extent of gene expression; the process is mitotically or meiotically heritable, or is stably self-propagated in the cytoplasm of a resting cell, and does not entail a change in DNA sequence.
macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
positive regulation of gene expression, epigenetic
Any epigenetic process that activates or increases the rate of gene expression.
positive regulation of biological process
Any process that activates or increases the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of macromolecule metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
all
This term is the most general term possible
positive regulation of metabolic process
Any process that activates or increases the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism.
regulation of metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism.
positive regulation of biological process
Any process that activates or increases the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
positive regulation of metabolic process
Any process that activates or increases the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism.
positive regulation of macromolecule metabolic process
Any process that increases the frequency, rate or extent of the chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
regulation of macromolecule metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
positive regulation of macromolecule metabolic process
Any process that increases the frequency, rate or extent of the chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
regulation of gene expression
Any process that modulates the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product or products (proteins or RNA). This includes the production of an RNA transcript as well as any processing to produce a mature RNA product or an mRNA (for protein-coding genes) and the translation of that mRNA into protein. Some protein processing events may be included when they are required to form an active form of a product from an inactive precursor form.
positive regulation of gene expression
Any process that increases the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product or products (proteins or RNA). This includes the production of an RNA transcript as well as any processing to produce a mature RNA product or an mRNA (for protein-coding genes) and the translation of that mRNA into protein. Some protein processing events may be included when they are required to form an active form of a product from an inactive precursor form.
positive regulation of gene expression
Any process that increases the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product or products (proteins or RNA). This includes the production of an RNA transcript as well as any processing to produce a mature RNA product or an mRNA (for protein-coding genes) and the translation of that mRNA into protein. Some protein processing events may be included when they are required to form an active form of a product from an inactive precursor form.
positive regulation of gene expression, epigenetic
Any epigenetic process that activates or increases the rate of gene expression.


ABCD1ATP-binding cassette, sub-family D (ALD), member 1 (205142_x_at), score: 0.71 ADCK2aarF domain containing kinase 2 (221893_s_at), score: 0.74 AGPAT11-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) (215535_s_at), score: 0.64 ALDH1L1aldehyde dehydrogenase 1 family, member L1 (215798_at), score: -0.67 ARSAarylsulfatase A (204443_at), score: 0.72 ATF5activating transcription factor 5 (204999_s_at), score: 0.62 ATN1atrophin 1 (40489_at), score: 0.81 B3GAT3beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) (203452_at), score: 0.79 BACE2beta-site APP-cleaving enzyme 2 (217867_x_at), score: 0.62 BAP1BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) (201419_at), score: 0.69 BAT2HLA-B associated transcript 2 (212081_x_at), score: 0.8 BAT2LHLA-B associated transcript 2-like (212068_s_at), score: 0.69 BTBD2BTB (POZ) domain containing 2 (207722_s_at), score: 0.74 C16orf57chromosome 16 open reading frame 57 (218060_s_at), score: 0.74 CABIN1calcineurin binding protein 1 (37652_at), score: 0.66 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (217849_s_at), score: 0.91 CDC42EP1CDC42 effector protein (Rho GTPase binding) 1 (204693_at), score: 0.69 CDC42EP4CDC42 effector protein (Rho GTPase binding) 4 (218062_x_at), score: 0.66 CENPTcentromere protein T (218148_at), score: 0.63 CICcapicua homolog (Drosophila) (212784_at), score: 0.84 CIZ1CDKN1A interacting zinc finger protein 1 (205516_x_at), score: 0.78 CLIP2CAP-GLY domain containing linker protein 2 (211031_s_at), score: 0.64 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: 0.79 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: 0.77 CScitrate synthase (208660_at), score: 0.65 DMWDdystrophia myotonica, WD repeat containing (33768_at), score: 0.64 DNM2dynamin 2 (202253_s_at), score: 0.61 DOK4docking protein 4 (209691_s_at), score: 0.65 DOPEY1dopey family member 1 (40612_at), score: -0.66 EHBP1L1EH domain binding protein 1-like 1 (221755_at), score: 0.61 EVLEnah/Vasp-like (217838_s_at), score: 0.63 FBXL14F-box and leucine-rich repeat protein 14 (213145_at), score: 0.64 FBXO17F-box protein 17 (220233_at), score: 0.67 FCF1FCF1 small subunit (SSU) processome component homolog (S. cerevisiae) (219927_at), score: -0.74 FHOD1formin homology 2 domain containing 1 (218530_at), score: 0.67 FLJ12529pre-mRNA cleavage factor I, 59 kDa subunit (217866_at), score: 0.79 FOSL1FOS-like antigen 1 (204420_at), score: 0.61 FOXK2forkhead box K2 (203064_s_at), score: 0.92 FURINfurin (paired basic amino acid cleaving enzyme) (201945_at), score: 0.67 GRINAglutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) (212090_at), score: 0.68 H1FXH1 histone family, member X (204805_s_at), score: 0.67 HAB1B1 for mucin (215778_x_at), score: -0.66 HMGA1high mobility group AT-hook 1 (210457_x_at), score: 0.71 HNRNPH2heterogeneous nuclear ribonucleoprotein H2 (H') (201132_at), score: -0.71 HNRNPUL1heterogeneous nuclear ribonucleoprotein U-like 1 (209675_s_at), score: 0.66 HSD17B6hydroxysteroid (17-beta) dehydrogenase 6 homolog (mouse) (37512_at), score: -0.66 HSF1heat shock transcription factor 1 (202344_at), score: 0.61 IDUAiduronidase, alpha-L- (205059_s_at), score: 0.85 JUNDjun D proto-oncogene (203751_x_at), score: 0.63 JUPjunction plakoglobin (201015_s_at), score: 0.68 KAL1Kallmann syndrome 1 sequence (205206_at), score: -0.66 LEPREL2leprecan-like 2 (204854_at), score: 0.78 LMF2lipase maturation factor 2 (212682_s_at), score: 0.7 LOC339047hypothetical protein LOC339047 (221501_x_at), score: 0.61 LOC90379hypothetical protein BC002926 (221849_s_at), score: 0.69 LRDDleucine-rich repeats and death domain containing (221640_s_at), score: 0.72 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: 1 MAP3K6mitogen-activated protein kinase kinase kinase 6 (219278_at), score: 0.81 MAP4microtubule-associated protein 4 (200836_s_at), score: 0.61 MAP7D1MAP7 domain containing 1 (217943_s_at), score: 0.74 MAST2microtubule associated serine/threonine kinase 2 (211593_s_at), score: 0.74 MED15mediator complex subunit 15 (222175_s_at), score: 0.63 MGAT1mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase (201126_s_at), score: 0.66 MINK1misshapen-like kinase 1 (zebrafish) (214246_x_at), score: 0.68 MLL4myeloid/lymphoid or mixed-lineage leukemia 4 (203419_at), score: 0.63 MUL1mitochondrial E3 ubiquitin ligase 1 (218246_at), score: 0.73 NBL1neuroblastoma, suppression of tumorigenicity 1 (37005_at), score: 0.72 NEFMneurofilament, medium polypeptide (205113_at), score: -0.69 NFATC4nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 (205897_at), score: 0.7 NPIPnuclear pore complex interacting protein (204538_x_at), score: 0.64 PARVBparvin, beta (37965_at), score: 0.8 PCDHG@protocadherin gamma cluster (215836_s_at), score: 0.74 PCDHGA1protocadherin gamma subfamily A, 1 (209079_x_at), score: 0.79 PIP5K1Cphosphatidylinositol-4-phosphate 5-kinase, type I, gamma (212518_at), score: 0.92 PKN1protein kinase N1 (202161_at), score: 0.73 PNPLA6patatin-like phospholipase domain containing 6 (203718_at), score: 0.66 POLR2Apolymerase (RNA) II (DNA directed) polypeptide A, 220kDa (202725_at), score: 0.8 POLSpolymerase (DNA directed) sigma (202466_at), score: 0.68 POM121POM121 membrane glycoprotein (rat) (212178_s_at), score: 0.75 POMZP3POM (POM121 homolog, rat) and ZP3 fusion (204148_s_at), score: 0.69 PRKACAprotein kinase, cAMP-dependent, catalytic, alpha (202801_at), score: 0.63 PRKCDprotein kinase C, delta (202545_at), score: 0.71 PTPN9protein tyrosine phosphatase, non-receptor type 9 (202958_at), score: 0.62 PTPROprotein tyrosine phosphatase, receptor type, O (211600_at), score: 0.63 RAD54L2RAD54-like 2 (S. cerevisiae) (213205_s_at), score: 0.7 RBM15BRNA binding motif protein 15B (202689_at), score: 0.71 REREarginine-glutamic acid dipeptide (RE) repeats (200940_s_at), score: 0.65 RNF220ring finger protein 220 (219988_s_at), score: 0.71 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: -0.91 RPRD2regulation of nuclear pre-mRNA domain containing 2 (212553_at), score: 0.69 RPS17P5ribosomal protein S17 pseudogene 5 (216348_at), score: -0.79 SBF1SET binding factor 1 (39835_at), score: 0.61 SBNO2strawberry notch homolog 2 (Drosophila) (204166_at), score: 0.7 SCAMP4secretory carrier membrane protein 4 (213244_at), score: 0.99 SDAD1SDA1 domain containing 1 (218607_s_at), score: 0.63 SF1splicing factor 1 (208313_s_at), score: 0.85 SF3A2splicing factor 3a, subunit 2, 66kDa (37462_i_at), score: 0.68 SMARCD1SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 (209518_at), score: 0.61 SMG6Smg-6 homolog, nonsense mediated mRNA decay factor (C. elegans) (214940_s_at), score: 0.71 SMG7Smg-7 homolog, nonsense mediated mRNA decay factor (C. elegans) (201794_s_at), score: 0.69 SMYD5SMYD family member 5 (209516_at), score: 0.65 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: 0.84 SPG7spastic paraplegia 7 (pure and complicated autosomal recessive) (202104_s_at), score: 0.64 SPRY4sprouty homolog 4 (Drosophila) (221489_s_at), score: 0.64 SSBP3single stranded DNA binding protein 3 (217991_x_at), score: 0.68 STRN4striatin, calmodulin binding protein 4 (217903_at), score: 0.78 TAF13TAF13 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 18kDa (205966_at), score: -0.66 TAPBPTAP binding protein (tapasin) (208829_at), score: 0.85 TBC1D10BTBC1 domain family, member 10B (220947_s_at), score: 0.63 TERF2telomeric repeat binding factor 2 (203611_at), score: 0.65 TIAF1TGFB1-induced anti-apoptotic factor 1 (202039_at), score: 0.63 TICAM2toll-like receptor adaptor molecule 2 (214658_at), score: -0.67 TMSB4Ythymosin beta 4, Y-linked (206769_at), score: -0.67 TSC2tuberous sclerosis 2 (215735_s_at), score: 0.64 TSKUtsukushin (218245_at), score: 0.69 TXLNAtaxilin alpha (212300_at), score: 0.75 UBTD1ubiquitin domain containing 1 (219172_at), score: 0.72 VEGFBvascular endothelial growth factor B (203683_s_at), score: 0.9 VPS37Bvacuolar protein sorting 37 homolog B (S. cerevisiae) (221704_s_at), score: 0.84 WDR42AWD repeat domain 42A (202249_s_at), score: 0.67 WDR6WD repeat domain 6 (217734_s_at), score: 0.7 XAB2XPA binding protein 2 (218110_at), score: 0.71 ZFP36L1zinc finger protein 36, C3H type-like 1 (211965_at), score: 0.7
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3407-raw-cel-1437949557.cel | 1 | 4 | Cockayne | hgu133a | CS | eGFP |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486011.cel | 19 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486171.cel | 27 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485971.cel | 17 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486031.cel | 20 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485991.cel | 18 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486071.cel | 22 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690344.cel | 10 | 5 | HGPS | hgu133a | none | GM00038C |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486411.cel | 39 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690304.cel | 8 | 5 | HGPS | hgu133a | none | GMO8398C |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |