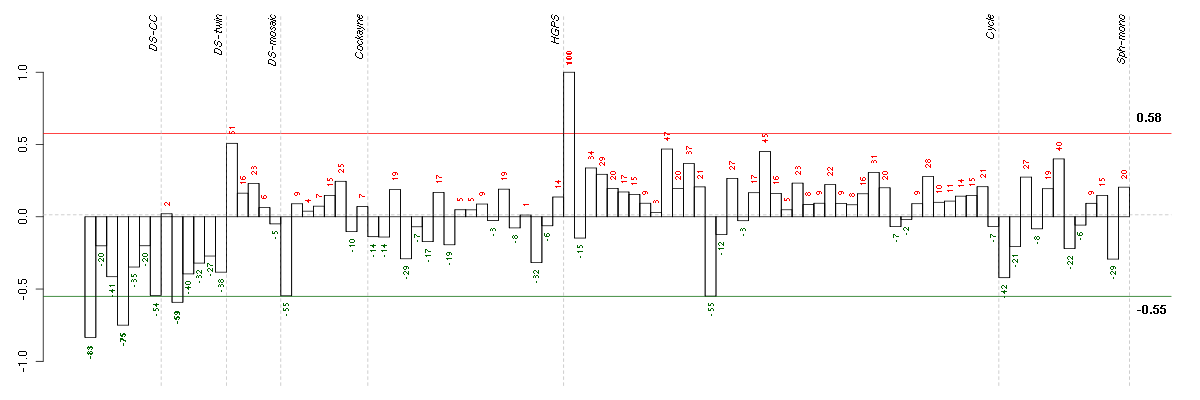

Under-expression is coded with green,
over-expression with red color.

metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
system process
A multicellular organismal process carried out by any of the organs or tissues in an organ system. An organ system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a biological objective.
signal transduction
The cascade of processes by which a signal interacts with a receptor, causing a change in the level or activity of a second messenger or other downstream target, and ultimately effecting a change in the functioning of the cell.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
The chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
regulation of nucleotide metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nucleotides.
cAMP biosynthetic process
The chemical reactions and pathways resulting in the formation of the nucleotide cAMP (cyclic AMP, adenosine 3',5'-cyclophosphate).
nucleoside phosphate metabolic process
The chemical reactions and pathways involving any phosphorylated nucleoside.
cell communication
Any process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
cell surface receptor linked signal transduction
Any series of molecular signals initiated by the binding of an extracellular ligand to a receptor on the surface of the target cell.
G-protein coupled receptor protein signaling pathway
The series of molecular signals generated as a consequence of a G-protein coupled receptor binding to its physiological ligand.
neurological system process
A organ system process carried out by any of the organs or tissues of neurological system.
sensory perception
The series of events required for an organism to receive a sensory stimulus, convert it to a molecular signal, and recognize and characterize the signal.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cAMP metabolic process
The chemical reactions and pathways involving the nucleotide cAMP (cyclic AMP, adenosine 3',5'-cyclophosphate).
biosynthetic process
The chemical reactions and pathways resulting in the formation of substances; typically the energy-requiring part of metabolism in which simpler substances are transformed into more complex ones.
nucleotide metabolic process
The chemical reactions and pathways involving a nucleotide, a nucleoside that is esterified with (ortho)phosphate or an oligophosphate at any hydroxyl group on the glycose moiety; may be mono-, di- or triphosphate; this definition includes cyclic nucleotides (nucleoside cyclic phosphates).
nucleotide biosynthetic process
The chemical reactions and pathways resulting in the formation of nucleotides, any nucleoside that is esterified with (ortho)phosphate or an oligophosphate at any hydroxyl group on the glycose moiety; may be mono-, di- or triphosphate; this definition includes cyclic-nucleotides (nucleoside cyclic phosphates).
cyclic nucleotide metabolic process
The chemical reactions and pathways involving a cyclic nucleotide, a nucleotide in which the phosphate group is in diester linkage to two positions on the sugar residue.
cyclic nucleotide biosynthetic process
The chemical reactions and pathways resulting in the formation of a cyclic nucleotide, a nucleotide in which the phosphate group is in diester linkage to two positions on the sugar residue.
regulation of biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
regulation of metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism.
regulation of cyclic nucleotide metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving cyclic nucleotides.
regulation of cyclic nucleotide biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of cyclic nucleotides.
regulation of nucleotide biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of nucleotides.
regulation of cAMP metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving the nucleotide cAMP (cyclic AMP, adenosine 3',5'-cyclophosphate).
regulation of cAMP biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of the nucleotide cAMP (cyclic AMP, adenosine 3',5'-cyclophosphate).
regulation of cyclase activity
Any process that modulates the frequency, rate or extent of cyclase activity.
regulation of cellular metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways by which individual cells transform chemical substances.
regulation of cellular biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
primary metabolic process
The chemical reactions and pathways involving those compounds which are formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
cellular biosynthetic process
The chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
regulation of adenylate cyclase activity
Any process that modulates the frequency, rate or extent of adenylate cyclase activity.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of catalytic activity
Any process that modulates the activity of an enzyme.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
cognition
The operation of the mind by which an organism becomes aware of objects of thought or perception; it includes the mental activities associated with thinking, learning, and memory.
regulation of lyase activity
Any process that modulates the frequency, rate or extent of lyase activity, the catalysis of the cleavage of C-C, C-O, C-N and other bonds by other means than by hydrolysis or oxidation, or conversely adding a group to a double bond. They differ from other enzymes in that two substrates are involved in one reaction direction, but only one in the other direction. When acting on the single substrate, a molecule is eliminated and this generates either a new double bond or a new ring.
nucleobase, nucleoside and nucleotide metabolic process
The chemical reactions and pathways involving nucleobases, nucleosides and nucleotides.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
regulation of molecular function
Any process that modulates the frequency, rate or extent of molecular functions. Molecular functions are elemental biological activities occurring at the molecular level, such as catalysis or binding.
all
This term is the most general term possible
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
regulation of metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances.
regulation of cellular metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways by which individual cells transform chemical substances.
cellular biosynthetic process
The chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
The chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
signal transduction
The cascade of processes by which a signal interacts with a receptor, causing a change in the level or activity of a second messenger or other downstream target, and ultimately effecting a change in the functioning of the cell.
regulation of cellular metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways by which individual cells transform chemical substances.
regulation of cellular biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
regulation of cellular biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
regulation of adenylate cyclase activity
Any process that modulates the frequency, rate or extent of adenylate cyclase activity.
regulation of nucleotide biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of nucleotides.
regulation of cyclase activity
Any process that modulates the frequency, rate or extent of cyclase activity.
regulation of cyclic nucleotide biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of cyclic nucleotides.
regulation of nucleotide metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nucleotides.
regulation of adenylate cyclase activity
Any process that modulates the frequency, rate or extent of adenylate cyclase activity.
regulation of cAMP biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of the nucleotide cAMP (cyclic AMP, adenosine 3',5'-cyclophosphate).
regulation of nucleotide biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of nucleotides.
cyclic nucleotide biosynthetic process
The chemical reactions and pathways resulting in the formation of a cyclic nucleotide, a nucleotide in which the phosphate group is in diester linkage to two positions on the sugar residue.
regulation of cyclic nucleotide metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving cyclic nucleotides.
regulation of cyclic nucleotide biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of cyclic nucleotides.
cAMP biosynthetic process
The chemical reactions and pathways resulting in the formation of the nucleotide cAMP (cyclic AMP, adenosine 3',5'-cyclophosphate).
regulation of cAMP metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving the nucleotide cAMP (cyclic AMP, adenosine 3',5'-cyclophosphate).
regulation of cAMP biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of the nucleotide cAMP (cyclic AMP, adenosine 3',5'-cyclophosphate).

plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
integral to membrane
Penetrating at least one phospholipid bilayer of a membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer. When used to describe a protein, indicates that all or part of the peptide sequence is embedded in the membrane.
extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.

peptide receptor activity
Combining with an extracellular or intracellular peptide to initiate a change in cell activity.
molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
signal transducer activity
Mediates the transfer of a signal from the outside to the inside of a cell by means other than the introduction of the signal molecule itself into the cell.
receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity.
transmembrane receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity, and spanning to the membrane of either the cell or an organelle.
G-protein coupled receptor activity
A receptor that binds an extracellular ligand and transmits the signal to a heterotrimeric G-protein complex. These receptors are characteristically seven-transmembrane receptors and are made up of hetero- or homodimers.
binding
The selective, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
neuropeptide receptor activity
Combining with a neuropeptide to initiate a change in cell activity.
peptide receptor activity, G-protein coupled
Combining with an extracellular or intracellular peptide to initiate a G-protein mediated change in cell activity. A G-protein is a signal transduction molecule that alternates between an inactive GDP-bound and an active GTP-bound state.
neurotransmitter receptor activity
Combining with a neurotransmitter to initiate a change in cell activity.
neurotransmitter binding
Interacting selectively with a neurotransmitter, any chemical substance that is capable of transmitting (or inhibiting the transmission of) a nerve impulse from a neuron to another cell.
peptide binding
Interacting selectively with peptides, any of a group of organic compounds comprising two or more amino acids linked by peptide bonds.
neuropeptide binding
Interacting selectively and stoichiometrically with neuropeptides, peptides with direct synaptic effects (peptide neurotransmitters) or indirect modulatory effects on the nervous system (peptide neuromodulators).
molecular transducer activity
The molecular function that accepts an input of one form and creates an output of a different form.
all
This term is the most general term possible
neuropeptide receptor activity
Combining with a neuropeptide to initiate a change in cell activity.
peptide receptor activity
Combining with an extracellular or intracellular peptide to initiate a change in cell activity.
neurotransmitter receptor activity
Combining with a neurotransmitter to initiate a change in cell activity.
neuropeptide receptor activity
Combining with a neuropeptide to initiate a change in cell activity.
peptide receptor activity, G-protein coupled
Combining with an extracellular or intracellular peptide to initiate a G-protein mediated change in cell activity. A G-protein is a signal transduction molecule that alternates between an inactive GDP-bound and an active GTP-bound state.
| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 04080 | 1.358e-02 | 1.625 | 8 | 78 | Neuroactive ligand-receptor interaction |
A1CFAPOBEC1 complementation factor (220951_s_at), score: -0.56 ABCG4ATP-binding cassette, sub-family G (WHITE), member 4 (207593_at), score: -0.65 ABOABO blood group (transferase A, alpha 1-3-N-acetylgalactosaminyltransferase; transferase B, alpha 1-3-galactosyltransferase) (216716_at), score: -0.76 ACSL6acyl-CoA synthetase long-chain family member 6 (211207_s_at), score: -0.82 ADAMDEC1ADAM-like, decysin 1 (206134_at), score: -0.8 ADAMTS9ADAM metallopeptidase with thrombospondin type 1 motif, 9 (220287_at), score: -0.81 ADAMTSL4ADAMTS-like 4 (220578_at), score: -0.54 ADRA2Aadrenergic, alpha-2A-, receptor (209869_at), score: -0.6 AGMATagmatine ureohydrolase (agmatinase) (219792_at), score: -0.54 AKR1D1aldo-keto reductase family 1, member D1 (delta 4-3-ketosteroid-5-beta-reductase) (207102_at), score: -0.57 APOMapolipoprotein M (205682_x_at), score: -0.58 ASAP3ArfGAP with SH3 domain, ankyrin repeat and PH domain 3 (219103_at), score: -0.55 AZGP1alpha-2-glycoprotein 1, zinc-binding (217014_s_at), score: -0.66 BEANbrain expressed, associated with Nedd4 (214068_at), score: -0.67 BMP10bone morphogenetic protein 10 (208292_at), score: -0.64 BRS3bombesin-like receptor 3 (207369_at), score: -0.79 BTNL3butyrophilin-like 3 (217207_s_at), score: -0.72 C15orf5chromosome 15 open reading frame 5 (208109_s_at), score: -0.67 C7orf28Achromosome 7 open reading frame 28A (201974_s_at), score: -0.64 C8orf60chromosome 8 open reading frame 60 (220712_at), score: -0.58 CA4carbonic anhydrase IV (206209_s_at), score: -0.58 CALCAcalcitonin-related polypeptide alpha (217561_at), score: -0.56 CATSPER2cation channel, sperm associated 2 (217588_at), score: -0.55 CCDC9coiled-coil domain containing 9 (206257_at), score: -0.74 CD28CD28 molecule (206545_at), score: -0.58 CDH19cadherin 19, type 2 (206898_at), score: -0.81 CEACAM5carcinoembryonic antigen-related cell adhesion molecule 5 (201884_at), score: -0.68 CEACAM6carcinoembryonic antigen-related cell adhesion molecule 6 (non-specific cross reacting antigen) (211657_at), score: -0.81 CHD2chromodomain helicase DNA binding protein 2 (203461_at), score: -0.53 CHN2chimerin (chimaerin) 2 (207486_x_at), score: -0.76 CHST4carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 4 (220446_s_at), score: -0.6 CLDN10claudin 10 (205328_at), score: -0.74 CLDN18claudin 18 (214135_at), score: -0.58 CLEC2DC-type lectin domain family 2, member D (220132_s_at), score: -0.58 CLEC4MC-type lectin domain family 4, member M (207995_s_at), score: -0.55 CNKSR2connector enhancer of kinase suppressor of Ras 2 (206731_at), score: -0.6 COL11A2collagen, type XI, alpha 2 (216993_s_at), score: -0.63 CR1complement component (3b/4b) receptor 1 (Knops blood group) (217484_at), score: -0.57 CSN2casein beta (207951_at), score: -0.56 CTTNBP2NLCTTNBP2 N-terminal like (214731_at), score: -0.63 CXCL10chemokine (C-X-C motif) ligand 10 (204533_at), score: -0.57 CXorf21chromosome X open reading frame 21 (220252_x_at), score: -0.56 CYP2E1cytochrome P450, family 2, subfamily E, polypeptide 1 (1431_at), score: -0.54 CYP3A4cytochrome P450, family 3, subfamily A, polypeptide 4 (205998_x_at), score: -0.6 DAOD-amino-acid oxidase (206878_at), score: -0.53 DESdesmin (216947_at), score: -0.64 DKK2dickkopf homolog 2 (Xenopus laevis) (219908_at), score: -0.66 DNASE1L3deoxyribonuclease I-like 3 (205554_s_at), score: -0.63 DRD5dopamine receptor D5 (208486_at), score: -0.62 DSC2desmocollin 2 (204751_x_at), score: -0.7 DTX4deltex homolog 4 (Drosophila) (212611_at), score: -0.6 DXS542X-linked retinopathy protein-like (222247_at), score: -0.62 ELNelastin (212670_at), score: -0.65 EPHA3EPH receptor A3 (211164_at), score: -0.56 ERBB3v-erb-b2 erythroblastic leukemia viral oncogene homolog 3 (avian) (202454_s_at), score: -0.54 ERBB4v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian) (214053_at), score: -0.63 FA2Hfatty acid 2-hydroxylase (219429_at), score: -0.57 FAM38Bfamily with sequence similarity 38, member B (219602_s_at), score: -0.61 FAM46Cfamily with sequence similarity 46, member C (220306_at), score: -0.54 FUT3fucosyltransferase 3 (galactoside 3(4)-L-fucosyltransferase, Lewis blood group) (214088_s_at), score: -0.56 GBA3glucosidase, beta, acid 3 (cytosolic) (219954_s_at), score: -0.58 GLRA1glycine receptor, alpha 1 (207972_at), score: -0.62 GNRHRgonadotropin-releasing hormone receptor (216341_s_at), score: -0.68 GP6glycoprotein VI (platelet) (220336_s_at), score: -0.63 GPATCH4G patch domain containing 4 (220596_at), score: -0.58 GPR12G protein-coupled receptor 12 (214558_at), score: -0.81 GPR63G protein-coupled receptor 63 (220993_s_at), score: -0.69 GRHL2grainyhead-like 2 (Drosophila) (219388_at), score: -0.56 HAVCR1hepatitis A virus cellular receptor 1 (207052_at), score: -0.57 HIST1H2ALhistone cluster 1, H2al (214554_at), score: -0.71 HIST1H4Ghistone cluster 1, H4g (208551_at), score: -0.55 HOXA7homeobox A7 (206848_at), score: -0.54 HOXD4homeobox D4 (205522_at), score: -0.55 HSPB6heat shock protein, alpha-crystallin-related, B6 (214767_s_at), score: -0.58 HUNKhormonally up-regulated Neu-associated kinase (219535_at), score: -0.53 ICA1islet cell autoantigen 1, 69kDa (211740_at), score: -0.75 IFNA5interferon, alpha 5 (214569_at), score: -0.55 KALRNkalirin, RhoGEF kinase (206078_at), score: -0.56 KCNIP1Kv channel interacting protein 1 (221307_at), score: -0.6 KCNJ16potassium inwardly-rectifying channel, subfamily J, member 16 (219564_at), score: -0.82 KIR3DX1killer cell immunoglobulin-like receptor, three domains, X1 (216428_x_at), score: -0.69 KLF1Kruppel-like factor 1 (erythroid) (210504_at), score: -0.53 KRT2keratin 2 (207908_at), score: -0.68 KRT8P12keratin 8 pseudogene 12 (222060_at), score: -0.55 LAX1lymphocyte transmembrane adaptor 1 (207734_at), score: -0.6 LHX3LIM homeobox 3 (221670_s_at), score: -0.82 LILRP2leukocyte immunoglobulin-like receptor pseudogene 2 (214561_at), score: -0.57 LOC100130703similar to hCG2042168 (207596_at), score: -0.57 LOC100132247similar to Uncharacterized protein KIAA0220 (215002_at), score: -0.65 LOC100188945cell division cycle associated 4 pseudogene (215109_at), score: -0.66 LOC286434hypothetical protein LOC286434 (222196_at), score: -0.54 LOC389906similar to Serine/threonine-protein kinase PRKX (Protein kinase PKX1) (59433_at), score: -0.73 LOC80054hypothetical LOC80054 (220465_at), score: -0.56 LOC93432maltase-glucoamylase-like pseudogene (216666_at), score: -0.75 LOH3CR2Aloss of heterozygosity, 3, chromosomal region 2, gene A (220244_at), score: -0.7 LONRF3LON peptidase N-terminal domain and ring finger 3 (220009_at), score: -0.67 MAP3K9mitogen-activated protein kinase kinase kinase 9 (213927_at), score: -0.56 MAPK8IP1mitogen-activated protein kinase 8 interacting protein 1 (213013_at), score: -0.61 MAPK8IP2mitogen-activated protein kinase 8 interacting protein 2 (205050_s_at), score: -0.6 MAPK8IP3mitogen-activated protein kinase 8 interacting protein 3 (213178_s_at), score: -0.57 MBmyoglobin (204179_at), score: -0.79 MEP1Bmeprin A, beta (207251_at), score: -0.64 MFNGMFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase (204153_s_at), score: -0.8 MGAT4Cmannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) (207447_s_at), score: -0.66 MGC5590hypothetical protein MGC5590 (220931_at), score: -0.63 MMRN2multimerin 2 (219091_s_at), score: -0.75 MOBPmyelin-associated oligodendrocyte basic protein (210193_at), score: -0.73 MSTP9macrophage stimulating, pseudogene 9 (213382_at), score: -0.66 MYBPC1myosin binding protein C, slow type (214087_s_at), score: -0.56 MYL4myosin, light chain 4, alkali; atrial, embryonic (210088_x_at), score: -0.59 MYO1Amyosin IA (211916_s_at), score: -0.91 MYO5Cmyosin VC (218966_at), score: -0.79 N4BP3Nedd4 binding protein 3 (214775_at), score: -0.73 NCRNA00092non-protein coding RNA 92 (215861_at), score: -0.65 NCRNA00093non-protein coding RNA 93 (210723_x_at), score: -0.55 NPY2Rneuropeptide Y receptor Y2 (210730_s_at), score: -0.56 NRN1neuritin 1 (218625_at), score: -0.86 NTNG1netrin G1 (206713_at), score: -0.53 NTRK2neurotrophic tyrosine kinase, receptor, type 2 (207152_at), score: -0.73 NTRK3neurotrophic tyrosine kinase, receptor, type 3 (217033_x_at), score: -0.58 NYXnyctalopin (221684_s_at), score: -0.58 OGFRopioid growth factor receptor (211512_s_at), score: -0.59 OR10C1olfactory receptor, family 10, subfamily C, member 1 (221339_at), score: -0.56 OR1E2olfactory receptor, family 1, subfamily E, member 2 (208587_s_at), score: -0.56 OR7E24olfactory receptor, family 7, subfamily E, member 24 (215463_at), score: -0.68 OVOL2ovo-like 2 (Drosophila) (211778_s_at), score: -0.57 PAX4paired box 4 (207867_at), score: -0.58 PDE3Aphosphodiesterase 3A, cGMP-inhibited (206388_at), score: -0.75 PDZRN4PDZ domain containing ring finger 4 (220595_at), score: -0.57 PECAM1platelet/endothelial cell adhesion molecule (208982_at), score: -0.82 PEG3paternally expressed 3 (209242_at), score: -0.65 PHF20L1PHD finger protein 20-like 1 (222133_s_at), score: -0.55 PLA2G7phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) (206214_at), score: -0.55 PLUNCpalate, lung and nasal epithelium associated (220542_s_at), score: -0.55 PMP2peripheral myelin protein 2 (206826_at), score: -0.55 PNPLA2patatin-like phospholipase domain containing 2 (212705_x_at), score: -0.7 PPIAL4Apeptidylprolyl isomerase A (cyclophilin A)-like 4A (217136_at), score: -0.59 PPP4R4protein phosphatase 4, regulatory subunit 4 (220672_at), score: -0.63 PROL1proline rich, lacrimal 1 (208004_at), score: -0.64 PROX1prospero homeobox 1 (207401_at), score: -0.62 PRSS7protease, serine, 7 (enterokinase) (217269_s_at), score: -0.9 PTPREprotein tyrosine phosphatase, receptor type, E (221840_at), score: -0.55 PYHIN1pyrin and HIN domain family, member 1 (216748_at), score: -0.77 RAPGEF4Rap guanine nucleotide exchange factor (GEF) 4 (205651_x_at), score: -0.6 RASL11BRAS-like, family 11, member B (219142_at), score: -0.55 RETret proto-oncogene (205879_x_at), score: -0.61 RIMS2regulating synaptic membrane exocytosis 2 (215478_at), score: -0.54 RNASE2ribonuclease, RNase A family, 2 (liver, eosinophil-derived neurotoxin) (206111_at), score: -0.66 RP11-35N6.1plasticity related gene 3 (219732_at), score: -0.82 RP3-377H14.5hypothetical LOC285830 (222279_at), score: -0.6 RP4-692D3.1hypothetical protein LOC728621 (221171_at), score: -0.59 RPGRIP1retinitis pigmentosa GTPase regulator interacting protein 1 (206608_s_at), score: -0.72 RRHretinal pigment epithelium-derived rhodopsin homolog (208314_at), score: -0.56 S100A14S100 calcium binding protein A14 (218677_at), score: -0.69 SCARF1scavenger receptor class F, member 1 (206995_x_at), score: -0.67 SCN10Asodium channel, voltage-gated, type X, alpha subunit (208578_at), score: -0.63 SFTPBsurfactant protein B (214354_x_at), score: -0.63 SH3TC2SH3 domain and tetratricopeptide repeats 2 (219710_at), score: -0.57 SLC10A1solute carrier family 10 (sodium/bile acid cotransporter family), member 1 (207185_at), score: -0.71 SLC10A2solute carrier family 10 (sodium/bile acid cotransporter family), member 2 (207095_at), score: -0.78 SLC12A3solute carrier family 12 (sodium/chloride transporters), member 3 (215274_at), score: -0.53 SLC15A1solute carrier family 15 (oligopeptide transporter), member 1 (211349_at), score: -0.68 SLC26A10solute carrier family 26, member 10 (214951_at), score: -0.57 SLC4A10solute carrier family 4, sodium bicarbonate transporter, member 10 (206830_at), score: -0.66 SLC6A14solute carrier family 6 (amino acid transporter), member 14 (219795_at), score: -0.67 SOCS3suppressor of cytokine signaling 3 (206359_at), score: -0.57 SRCAPSnf2-related CREBBP activator protein (212275_s_at), score: -0.57 SYN2synapsin II (210247_at), score: -0.59 TAAR2trace amine associated receptor 2 (221394_at), score: -0.78 TACR1tachykinin receptor 1 (208048_at), score: -0.79 TAS2R9taste receptor, type 2, member 9 (221461_at), score: -0.68 TBX21T-box 21 (220684_at), score: -0.63 TBX6T-box 6 (207684_at), score: -0.59 TGM4transglutaminase 4 (prostate) (217566_s_at), score: -0.55 TLR1toll-like receptor 1 (210176_at), score: -0.71 TMEM92transmembrane protein 92 (216791_at), score: -0.57 TMSB4Ythymosin beta 4, Y-linked (206769_at), score: -0.55 TP63tumor protein p63 (209863_s_at), score: -0.65 TRA@T cell receptor alpha locus (216540_at), score: -0.72 TSPY1testis specific protein, Y-linked 1 (216374_at), score: -0.71 TULP2tubby like protein 2 (206733_at), score: -0.53 VGFVGF nerve growth factor inducible (205586_x_at), score: -1 ZNF117zinc finger protein 117 (207605_x_at), score: -0.55
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| ctrl a 08-03.CEL | 1 | 1 | DS-CC | hgu133a | none | DS-CC 1 |
| t21a 08-03.CEL | 4 | 1 | DS-CC | hgu133a | Down | DS-CC 4 |
| 2Twin.CEL | 2 | 2 | DS-twin | hgu133plus2 | none | DS-twin 2 |
| E-TABM-263-raw-cel-1515485911.cel | 14 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485651.cel | 1 | 6 | Cycle | hgu133a2 | none | Cycle 1 |