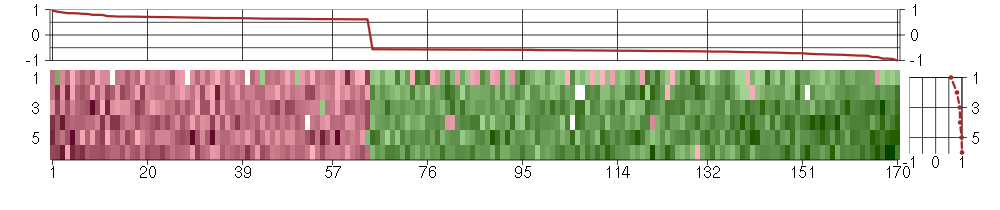

Under-expression is coded with green,
over-expression with red color.



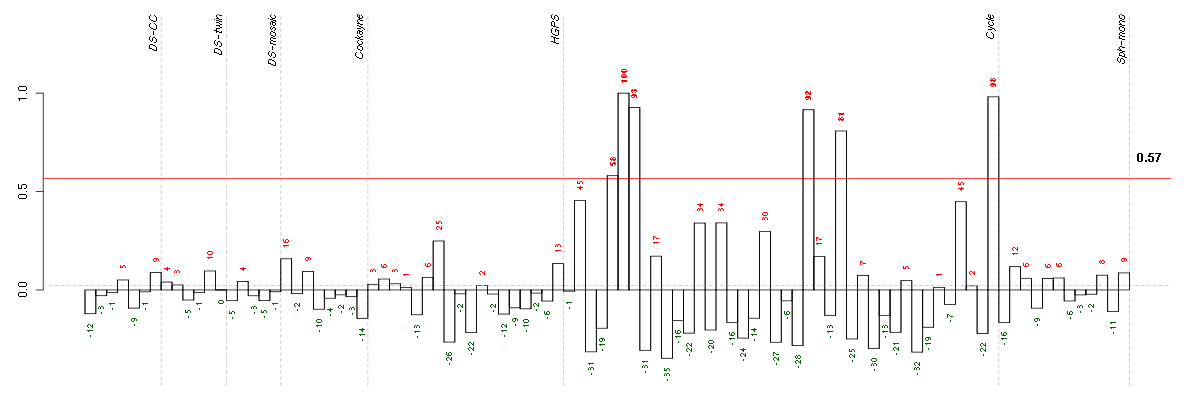
ABLIM1actin binding LIM protein 1 (200965_s_at), score: -0.63 ACTC1actin, alpha, cardiac muscle 1 (205132_at), score: 0.62 ADCY3adenylate cyclase 3 (209321_s_at), score: -0.93 ARL17P1ADP-ribosylation factor-like 17 pseudogene 1 (210718_s_at), score: -0.57 ATMataxia telangiectasia mutated (210858_x_at), score: 0.62 ATP6V0E1ATPase, H+ transporting, lysosomal 9kDa, V0 subunit e1 (214149_s_at), score: -0.64 BIN1bridging integrator 1 (210201_x_at), score: -0.71 BRCA2breast cancer 2, early onset (208368_s_at), score: 0.79 BRIP1BRCA1 interacting protein C-terminal helicase 1 (221703_at), score: 0.62 BTBD2BTB (POZ) domain containing 2 (207722_s_at), score: -0.61 C14orf106chromosome 14 open reading frame 106 (206500_s_at), score: 0.83 C16orf57chromosome 16 open reading frame 57 (218060_s_at), score: -0.56 C19orf40chromosome 19 open reading frame 40 (214816_x_at), score: 0.68 C1orf38chromosome 1 open reading frame 38 (210785_s_at), score: -0.66 C4orf15chromosome 4 open reading frame 15 (210054_at), score: 0.63 C5orf28chromosome 5 open reading frame 28 (219029_at), score: 0.64 CASC3cancer susceptibility candidate 3 (207842_s_at), score: -0.58 CASP2caspase 2, apoptosis-related cysteine peptidase (209811_at), score: 0.61 CCDC101coiled-coil domain containing 101 (48117_at), score: 0.66 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (217849_s_at), score: -0.63 CDR2Lcerebellar degeneration-related protein 2-like (213230_at), score: -0.61 CENPC1centromere protein C 1 (204739_at), score: 0.68 CENPJcentromere protein J (220885_s_at), score: 0.72 CENPQcentromere protein Q (219294_at), score: 0.71 CEP192centrosomal protein 192kDa (218827_s_at), score: 0.63 CEP57centrosomal protein 57kDa (203491_s_at), score: 0.72 CHST2carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2 (203921_at), score: -0.81 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: -0.57 COL10A1collagen, type X, alpha 1 (217428_s_at), score: -0.79 COL18A1collagen, type XVIII, alpha 1 (209081_s_at), score: -0.61 CORO2Bcoronin, actin binding protein, 2B (209789_at), score: -0.6 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: -0.72 CTPS2CTP synthase II (219080_s_at), score: 0.61 CUX1cut-like homeobox 1 (214743_at), score: 0.62 CYP7B1cytochrome P450, family 7, subfamily B, polypeptide 1 (207386_at), score: -0.65 DDX54DEAD (Asp-Glu-Ala-Asp) box polypeptide 54 (219111_s_at), score: -0.58 DENND4BDENN/MADD domain containing 4B (202860_at), score: -0.67 DESdesmin (216947_at), score: -0.63 DLG4discs, large homolog 4 (Drosophila) (204592_at), score: -0.64 DLSTPdihydrolipoamide S-succinyltransferase pseudogene (E2 component of 2-oxo-glutarate complex) (215210_s_at), score: 0.67 DMWDdystrophia myotonica, WD repeat containing (33768_at), score: -0.87 DNM3dynamin 3 (209839_at), score: -0.63 DSCR6Down syndrome critical region gene 6 (207267_s_at), score: -0.57 E2F8E2F transcription factor 8 (219990_at), score: 0.7 ELL3elongation factor RNA polymerase II-like 3 (219517_at), score: -0.62 EML2echinoderm microtubule associated protein like 2 (204398_s_at), score: 0.71 EPHA4EPH receptor A4 (206114_at), score: 0.72 F11coagulation factor XI (206610_s_at), score: 0.64 FAM131Afamily with sequence similarity 131, member A (221904_at), score: -0.65 FAM168Bfamily with sequence similarity 168, member B (212017_at), score: -0.56 FASNfatty acid synthase (212218_s_at), score: -0.68 FBXO17F-box protein 17 (220233_at), score: -0.57 FKBPLFK506 binding protein like (219187_at), score: 0.69 FKRPfukutin related protein (219853_at), score: -0.69 FLJ12529pre-mRNA cleavage factor I, 59 kDa subunit (217866_at), score: -0.75 FLJ20674hypothetical protein FLJ20674 (220137_at), score: 0.66 FOXK2forkhead box K2 (203064_s_at), score: -1 GFPT2glutamine-fructose-6-phosphate transaminase 2 (205100_at), score: -0.58 GMCL1germ cell-less homolog 1 (Drosophila) (218458_at), score: 0.66 GOLPH3golgi phosphoprotein 3 (coat-protein) (217803_at), score: -0.94 GP1BAglycoprotein Ib (platelet), alpha polypeptide (207389_at), score: 0.62 GRIK2glutamate receptor, ionotropic, kainate 2 (213845_at), score: 0.64 H2BFSH2B histone family, member S (208579_x_at), score: 0.67 HGFhepatocyte growth factor (hepapoietin A; scatter factor) (209960_at), score: -0.56 HIST1H2BHhistone cluster 1, H2bh (208546_x_at), score: 0.72 HNRNPA3P1heterogeneous nuclear ribonucleoprotein A3 pseudogene 1 (206809_s_at), score: -0.6 HSD17B6hydroxysteroid (17-beta) dehydrogenase 6 homolog (mouse) (37512_at), score: 0.85 HSPA12Aheat shock 70kDa protein 12A (214434_at), score: -0.74 IFI44interferon-induced protein 44 (214453_s_at), score: -0.6 INSIG1insulin induced gene 1 (201627_s_at), score: -0.63 ITGA1integrin, alpha 1 (214660_at), score: 0.62 JARID1Cjumonji, AT rich interactive domain 1C (202383_at), score: -0.62 KATNB1katanin p80 (WD repeat containing) subunit B 1 (203162_s_at), score: 0.74 KCNN2potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2 (220116_at), score: 0.82 KCTD13potassium channel tetramerisation domain containing 13 (45653_at), score: -0.58 KIAA0317KIAA0317 (202128_at), score: -0.65 KIAA1009KIAA1009 (206005_s_at), score: 0.86 KIAA1305KIAA1305 (220911_s_at), score: -0.69 KIF1Bkinesin family member 1B (209234_at), score: -0.58 KRT7keratin 7 (209016_s_at), score: 0.7 LOC100132540similar to LOC339047 protein (214870_x_at), score: -0.6 LOC100133105hypothetical protein LOC100133105 (214237_x_at), score: 0.67 LOC128192similar to peptidyl-Pro cis trans isomerase (217346_at), score: 0.65 LOC339047hypothetical protein LOC339047 (221501_x_at), score: -0.63 LRDDleucine-rich repeats and death domain containing (221640_s_at), score: -0.57 LRIG1leucine-rich repeats and immunoglobulin-like domains 1 (211596_s_at), score: -0.62 LRP8low density lipoprotein receptor-related protein 8, apolipoprotein e receptor (205282_at), score: 0.67 LY75lymphocyte antigen 75 (205668_at), score: -0.64 MAP2K3mitogen-activated protein kinase kinase 3 (207667_s_at), score: -0.65 MAPKAPK3mitogen-activated protein kinase-activated protein kinase 3 (202788_at), score: -0.59 MAST4microtubule associated serine/threonine kinase family member 4 (40016_g_at), score: -0.78 MC4Rmelanocortin 4 receptor (221467_at), score: 0.96 MEF2Bmyocyte enhancer factor 2B (209926_at), score: 0.63 MEF2Cmyocyte enhancer factor 2C (209199_s_at), score: -0.57 MINK1misshapen-like kinase 1 (zebrafish) (214246_x_at), score: -0.57 MLL4myeloid/lymphoid or mixed-lineage leukemia 4 (203419_at), score: -0.61 MOCOSmolybdenum cofactor sulfurase (219959_at), score: 0.62 NCKAP1NCK-associated protein 1 (207738_s_at), score: 0.64 NDUFA2NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2, 8kDa (213550_s_at), score: -0.64 NEFMneurofilament, medium polypeptide (205113_at), score: 0.79 NFATC4nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 (205897_at), score: -0.57 NPATnuclear protein, ataxia-telangiectasia locus (209798_at), score: 0.64 NUDT11nudix (nucleoside diphosphate linked moiety X)-type motif 11 (219855_at), score: -0.77 NUPL2nucleoporin like 2 (204003_s_at), score: 0.74 OAS22'-5'-oligoadenylate synthetase 2, 69/71kDa (204972_at), score: -0.62 PCDHGA1protocadherin gamma subfamily A, 1 (209079_x_at), score: -0.6 PHC1polyhomeotic homolog 1 (Drosophila) (218338_at), score: -0.76 PHF14PHD finger protein 14 (204525_at), score: -0.58 PIGLphosphatidylinositol glycan anchor biosynthesis, class L (213889_at), score: -0.67 PKNOX1PBX/knotted 1 homeobox 1 (221883_at), score: 0.64 PLCE1phospholipase C, epsilon 1 (205112_at), score: 0.72 PMS2PMS2 postmeiotic segregation increased 2 (S. cerevisiae) (209805_at), score: 0.68 POLHpolymerase (DNA directed), eta (219380_x_at), score: 0.71 POLSpolymerase (DNA directed) sigma (202466_at), score: -0.57 POM121POM121 membrane glycoprotein (rat) (212178_s_at), score: -0.71 PRKG2protein kinase, cGMP-dependent, type II (207505_at), score: -0.57 PTPN9protein tyrosine phosphatase, non-receptor type 9 (202958_at), score: -0.7 PXNpaxillin (211823_s_at), score: -0.56 RAB22ARAB22A, member RAS oncogene family (218360_at), score: -0.73 RAD54L2RAD54-like 2 (S. cerevisiae) (213205_s_at), score: -0.81 RBM15BRNA binding motif protein 15B (202689_at), score: -0.57 REREarginine-glutamic acid dipeptide (RE) repeats (200940_s_at), score: -0.67 RNF187ring finger protein 187 (212155_at), score: -0.65 RNF220ring finger protein 220 (219988_s_at), score: -0.62 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: 0.69 RPS11ribosomal protein S11 (213350_at), score: -0.58 RPS17P5ribosomal protein S17 pseudogene 5 (216348_at), score: 0.7 SCAMP4secretory carrier membrane protein 4 (213244_at), score: -0.58 SCN9Asodium channel, voltage-gated, type IX, alpha subunit (206950_at), score: 0.65 SETBP1SET binding protein 1 (205933_at), score: -0.69 SF1splicing factor 1 (208313_s_at), score: -0.6 SH2B3SH2B adaptor protein 3 (203320_at), score: -0.58 SH2D3ASH2 domain containing 3A (222169_x_at), score: 0.62 SHQ1SHQ1 homolog (S. cerevisiae) (63009_at), score: -0.71 SIRT6sirtuin (silent mating type information regulation 2 homolog) 6 (S. cerevisiae) (219613_s_at), score: -0.86 SMG7Smg-7 homolog, nonsense mediated mRNA decay factor (C. elegans) (201794_s_at), score: -0.68 SNX27sorting nexin family member 27 (221006_s_at), score: -0.75 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: -0.81 SSBP3single stranded DNA binding protein 3 (217991_x_at), score: -0.56 ST3GAL5ST3 beta-galactoside alpha-2,3-sialyltransferase 5 (203217_s_at), score: -0.63 ST7suppression of tumorigenicity 7 (207871_s_at), score: 0.64 STAU1staufen, RNA binding protein, homolog 1 (Drosophila) (207320_x_at), score: -0.57 STAU2staufen, RNA binding protein, homolog 2 (Drosophila) (204226_at), score: 0.66 STK3serine/threonine kinase 3 (STE20 homolog, yeast) (204068_at), score: 0.69 TAPBPTAP binding protein (tapasin) (208829_at), score: -0.58 TBX2T-box 2 (40560_at), score: -0.56 TBX3T-box 3 (219682_s_at), score: -0.79 TLE2transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila) (40837_at), score: -0.59 TMEM149transmembrane protein 149 (219690_at), score: 0.91 TNFRSF10Ctumor necrosis factor receptor superfamily, member 10c, decoy without an intracellular domain (206222_at), score: -0.61 TNFRSF25tumor necrosis factor receptor superfamily, member 25 (219423_x_at), score: -0.56 TNIP1TNFAIP3 interacting protein 1 (207196_s_at), score: -0.61 TOP3Atopoisomerase (DNA) III alpha (214300_s_at), score: 0.89 TSKUtsukushin (218245_at), score: -0.93 TTC13tetratricopeptide repeat domain 13 (219481_at), score: 0.7 TTLL7tubulin tyrosine ligase-like family, member 7 (219882_at), score: 0.62 TXLNAtaxilin alpha (212300_at), score: -0.61 UBTD1ubiquitin domain containing 1 (219172_at), score: -0.58 VPS37Bvacuolar protein sorting 37 homolog B (S. cerevisiae) (221704_s_at), score: -0.66 WDR42AWD repeat domain 42A (202249_s_at), score: -0.64 WDR6WD repeat domain 6 (217734_s_at), score: -0.77 WNT2Bwingless-type MMTV integration site family, member 2B (206459_s_at), score: 0.63 YPEL1yippee-like 1 (Drosophila) (206063_x_at), score: 0.79 ZDHHC14zinc finger, DHHC-type containing 14 (219247_s_at), score: 0.86 ZFP106zinc finger protein 106 homolog (mouse) (217781_s_at), score: -0.56 ZNF365zinc finger protein 365 (206448_at), score: -0.62 ZNF529zinc finger protein 529 (215307_at), score: 0.67 ZNF580zinc finger protein 580 (220748_s_at), score: -0.57 ZNF702Pzinc finger protein 702 pseudogene (206557_at), score: -0.56 ZNF814zinc finger protein 814 (60794_f_at), score: -0.59
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485731.cel | 5 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486151.cel | 26 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |