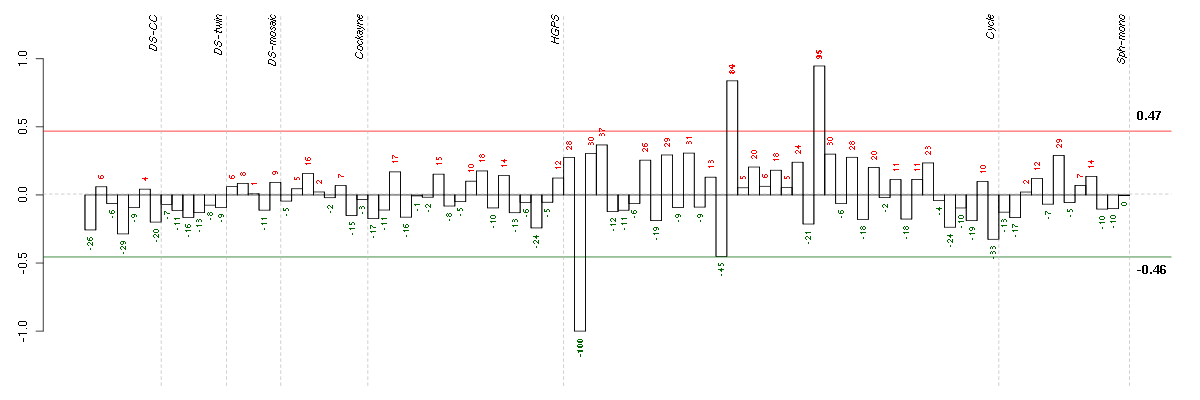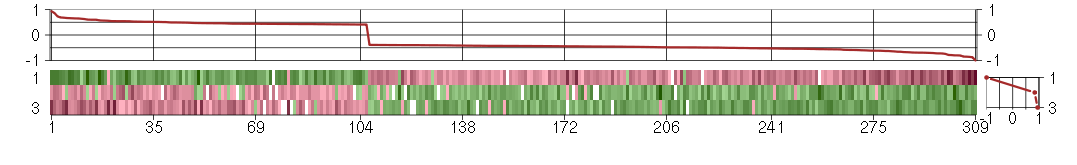

Under-expression is coded with green,
over-expression with red color.


extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
all
This term is the most general term possible

molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
signal transducer activity
Mediates the transfer of a signal from the outside to the inside of a cell by means other than the introduction of the signal molecule itself into the cell.
receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity.
transmembrane receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity, and spanning to the membrane of either the cell or an organelle.
molecular transducer activity
The molecular function that accepts an input of one form and creates an output of a different form.
all
This term is the most general term possible
A4GALTalpha 1,4-galactosyltransferase (219488_at), score: -0.45 ABCA7ATP-binding cassette, sub-family A (ABC1), member 7 (219577_s_at), score: -1 ACSS3acyl-CoA synthetase short-chain family member 3 (219616_at), score: 0.43 ADAMTS7ADAM metallopeptidase with thrombospondin type 1 motif, 7 (220705_s_at), score: -0.6 ADAMTSL4ADAMTS-like 4 (220578_at), score: 0.44 ADORA1adenosine A1 receptor (216220_s_at), score: -0.47 ADRA1Dadrenergic, alpha-1D-, receptor (210961_s_at), score: -0.69 ALDH1L1aldehyde dehydrogenase 1 family, member L1 (215798_at), score: -0.45 ALDH3A1aldehyde dehydrogenase 3 family, memberA1 (205623_at), score: -0.51 ALPPL2alkaline phosphatase, placental-like 2 (216377_x_at), score: -0.49 ALS2CR8amyotrophic lateral sclerosis 2 (juvenile) chromosome region, candidate 8 (219834_at), score: 0.46 AMELXamelogenin (amelogenesis imperfecta 1, X-linked) (208410_x_at), score: -0.45 ANGEL1angel homolog 1 (Drosophila) (36865_at), score: -0.43 ANKRD46ankyrin repeat domain 46 (212731_at), score: 0.43 AP1G2adaptor-related protein complex 1, gamma 2 subunit (201613_s_at), score: -0.43 APOC4apolipoprotein C-IV (206738_at), score: -0.51 APOL1apolipoprotein L, 1 (209546_s_at), score: -0.59 ARHGEF15Rho guanine nucleotide exchange factor (GEF) 15 (217348_x_at), score: -0.48 ASB9ankyrin repeat and SOCS box-containing 9 (205673_s_at), score: 0.44 ASPHD1aspartate beta-hydroxylase domain containing 1 (214993_at), score: -0.78 ATHL1ATH1, acid trehalase-like 1 (yeast) (219359_at), score: -0.67 ATMataxia telangiectasia mutated (210858_x_at), score: -0.43 ATP11AATPase, class VI, type 11A (215842_s_at), score: -0.4 ATP2C2ATPase, Ca++ transporting, type 2C, member 2 (214798_at), score: -0.47 ATP7BATPase, Cu++ transporting, beta polypeptide (204624_at), score: 0.46 ATXN2Lataxin 2-like (201806_s_at), score: -0.47 AZGP1alpha-2-glycoprotein 1, zinc-binding (217014_s_at), score: -0.4 BAHD1bromo adjacent homology domain containing 1 (203051_at), score: -0.42 BCAMbasal cell adhesion molecule (Lutheran blood group) (40093_at), score: -0.63 BEANbrain expressed, associated with Nedd4 (214068_at), score: 0.43 BEND5BEN domain containing 5 (219670_at), score: -0.49 BMP10bone morphogenetic protein 10 (208292_at), score: -0.43 BRAFv-raf murine sarcoma viral oncogene homolog B1 (206044_s_at), score: 0.68 BTF3L2basic transcription factor 3, like 2 (217461_x_at), score: -0.45 C10orf84chromosome 10 open reading frame 84 (218390_s_at), score: 0.54 C18orf1chromosome 18 open reading frame 1 (209574_s_at), score: 0.47 C1orf69chromosome 1 open reading frame 69 (215490_at), score: -0.51 C1QTNF1C1q and tumor necrosis factor related protein 1 (220975_s_at), score: -0.43 C21orf2chromosome 21 open reading frame 2 (203996_s_at), score: -0.44 C2orf37chromosome 2 open reading frame 37 (220172_at), score: 0.54 C2orf56chromosome 2 open reading frame 56 (220943_s_at), score: 0.52 C4Acomplement component 4A (Rodgers blood group) (214428_x_at), score: -0.41 C6orf35chromosome 6 open reading frame 35 (218453_s_at), score: 0.41 C7orf28Achromosome 7 open reading frame 28A (201974_s_at), score: -0.43 CA4carbonic anhydrase IV (206209_s_at), score: -0.44 CASP7caspase 7, apoptosis-related cysteine peptidase (207181_s_at), score: -0.39 CASP8caspase 8, apoptosis-related cysteine peptidase (213373_s_at), score: 0.42 CASQ1calsequestrin 1 (fast-twitch, skeletal muscle) (219645_at), score: 0.54 CATSPER2cation channel, sperm associated 2 (217588_at), score: -0.6 CC2D1Acoiled-coil and C2 domain containing 1A (58994_at), score: -0.54 CCDC40coiled-coil domain containing 40 (220592_at), score: -0.67 CCDC68coiled-coil domain containing 68 (220180_at), score: 0.5 CCDC9coiled-coil domain containing 9 (206257_at), score: -0.5 CCDC94coiled-coil domain containing 94 (204335_at), score: -0.43 CD22CD22 molecule (38521_at), score: -0.4 CD79BCD79b molecule, immunoglobulin-associated beta (205297_s_at), score: -0.4 CD80CD80 molecule (207176_s_at), score: 0.44 CDADC1cytidine and dCMP deaminase domain containing 1 (221015_s_at), score: 0.42 CEP72centrosomal protein 72kDa (219531_at), score: 0.43 CFHR2complement factor H-related 2 (206910_x_at), score: -0.49 CHMLchoroideremia-like (Rab escort protein 2) (206079_at), score: 0.44 CLCN2chloride channel 2 (213499_at), score: -0.43 CNTLNcentlein, centrosomal protein (220095_at), score: 0.52 CRLF1cytokine receptor-like factor 1 (206315_at), score: -0.44 CROCCciliary rootlet coiled-coil, rootletin (206274_s_at), score: -0.43 CTAGE1cutaneous T-cell lymphoma-associated antigen 1 (220957_at), score: 0.65 CYorf14chromosome Y open reading frame 14 (207063_at), score: 0.49 CYorf15Bchromosome Y open reading frame 15B (214131_at), score: -0.62 CYTH4cytohesin 4 (219183_s_at), score: -0.56 DCAKDdephospho-CoA kinase domain containing (221224_s_at), score: -0.54 DCLRE1BDNA cross-link repair 1B (PSO2 homolog, S. cerevisiae) (219490_s_at), score: 0.44 DHX32DEAH (Asp-Glu-Ala-His) box polypeptide 32 (218198_at), score: 0.42 DKFZp434H1419hypothetical protein DKFZp434H1419 (214717_at), score: -0.4 DMBT1deleted in malignant brain tumors 1 (208250_s_at), score: -0.85 DNA2DNA replication helicase 2 homolog (yeast) (213647_at), score: 0.43 DOHHdeoxyhypusine hydroxylase/monooxygenase (208141_s_at), score: -0.4 DSPPdentin sialophosphoprotein (221681_s_at), score: -0.47 DTNAdystrobrevin, alpha (205741_s_at), score: -0.44 DXS542X-linked retinopathy protein-like (222247_at), score: -0.53 EIF2C4eukaryotic translation initiation factor 2C, 4 (219190_s_at), score: 0.46 ELAC1elaC homolog 1 (E. coli) (219325_s_at), score: -0.39 ELK4ELK4, ETS-domain protein (SRF accessory protein 1) (206919_at), score: 0.51 ELLelongation factor RNA polymerase II (204095_s_at), score: -0.45 ENO3enolase 3 (beta, muscle) (204483_at), score: -0.46 EPHB4EPH receptor B4 (216680_s_at), score: -0.39 EPHB6EPH receptor B6 (204718_at), score: -0.41 EPN1epsin 1 (221141_x_at), score: -0.74 EPS8L1EPS8-like 1 (218778_x_at), score: -0.54 ETAA1Ewing tumor-associated antigen 1 (219216_at), score: 0.42 EXOC2exocyst complex component 2 (219349_s_at), score: 0.49 EXPH5exophilin 5 (214734_at), score: 0.73 F11coagulation factor XI (206610_s_at), score: -0.55 FAM59Afamily with sequence similarity 59, member A (219377_at), score: 0.43 FCER1GFc fragment of IgE, high affinity I, receptor for; gamma polypeptide (204232_at), score: -0.46 FGF13fibroblast growth factor 13 (205110_s_at), score: 0.44 FKBP10FK506 binding protein 10, 65 kDa (219249_s_at), score: -0.79 FLCNfolliculin (215645_at), score: -0.44 FLRT3fibronectin leucine rich transmembrane protein 3 (219250_s_at), score: 0.55 FLT3LGfms-related tyrosine kinase 3 ligand (210607_at), score: -0.58 FUT3fucosyltransferase 3 (galactoside 3(4)-L-fucosyltransferase, Lewis blood group) (214088_s_at), score: -0.52 FZD3frizzled homolog 3 (Drosophila) (219683_at), score: -0.5 GAB1GRB2-associated binding protein 1 (207112_s_at), score: -0.45 GABRR2gamma-aminobutyric acid (GABA) receptor, rho 2 (208217_at), score: -0.45 GARNL1GTPase activating Rap/RanGAP domain-like 1 (214855_s_at), score: 0.46 GASTgastrin (208138_at), score: -0.53 GATMglycine amidinotransferase (L-arginine:glycine amidinotransferase) (203178_at), score: 0.66 GBA3glucosidase, beta, acid 3 (cytosolic) (219954_s_at), score: -0.52 GCHFRGTP cyclohydrolase I feedback regulator (204867_at), score: -0.4 GGT1gamma-glutamyltransferase 1 (209919_x_at), score: -0.51 GIPC2GIPC PDZ domain containing family, member 2 (219970_at), score: 0.42 GLCEglucuronic acid epimerase (213552_at), score: 0.42 GNALguanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type (206355_at), score: 0.47 GNAO1guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O (204762_s_at), score: 0.48 GNL1guanine nucleotide binding protein-like 1 (203307_at), score: -0.45 GPR144G protein-coupled receptor 144 (216289_at), score: -0.8 GSDMDgasdermin D (218154_at), score: -0.4 GTF2H4general transcription factor IIH, polypeptide 4, 52kDa (203577_at), score: -0.41 GVIN1GTPase, very large interferon inducible 1 (220577_at), score: -0.4 GZMMgranzyme M (lymphocyte met-ase 1) (207460_at), score: -0.39 HAB1B1 for mucin (215778_x_at), score: -0.51 HAMPhepcidin antimicrobial peptide (220491_at), score: -0.53 HAO2hydroxyacid oxidase 2 (long chain) (220801_s_at), score: -0.62 HCG8HLA complex group 8 (215985_at), score: 0.46 HCLS1hematopoietic cell-specific Lyn substrate 1 (202957_at), score: 0.56 HIPK2homeodomain interacting protein kinase 2 (219028_at), score: -0.49 HIST1H2AGhistone cluster 1, H2ag (207156_at), score: 0.54 HIST1H2BChistone cluster 1, H2bc (214455_at), score: 0.49 HIST1H4Ghistone cluster 1, H4g (208551_at), score: -0.44 HIST1H4Hhistone cluster 1, H4h (208180_s_at), score: 0.41 HLA-DRB1major histocompatibility complex, class II, DR beta 1 (209312_x_at), score: -0.42 HPNhepsin (204934_s_at), score: -0.44 HSPA4Lheat shock 70kDa protein 4-like (205543_at), score: 0.47 HSPA6heat shock 70kDa protein 6 (HSP70B') (117_at), score: -0.69 ICAM2intercellular adhesion molecule 2 (204683_at), score: -0.53 IDI2isopentenyl-diphosphate delta isomerase 2 (217631_at), score: -0.4 IGHG1immunoglobulin heavy constant gamma 1 (G1m marker) (211693_at), score: -0.85 IGLL3immunoglobulin lambda-like polypeptide 3 (215946_x_at), score: -0.55 IL22RA1interleukin 22 receptor, alpha 1 (220056_at), score: -0.43 IL23Ainterleukin 23, alpha subunit p19 (211796_s_at), score: -0.69 IL9Rinterleukin 9 receptor (217212_s_at), score: -0.45 INE1inactivation escape 1 (non-protein coding) (207252_at), score: -0.49 IPPintracisternal A particle-promoted polypeptide (219843_at), score: 0.45 ISOC2isochorismatase domain containing 2 (218893_at), score: -0.41 ITGALintegrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide) (213475_s_at), score: -0.49 ITPR1inositol 1,4,5-triphosphate receptor, type 1 (203710_at), score: 0.67 KAT2BK(lysine) acetyltransferase 2B (203845_at), score: 0.42 KCNJ15potassium inwardly-rectifying channel, subfamily J, member 15 (210119_at), score: 0.51 KCTD17potassium channel tetramerisation domain containing 17 (205561_at), score: -0.63 KERAkeratocan (220504_at), score: 0.53 KIF16Bkinesin family member 16B (219570_at), score: 0.54 KIF26Bkinesin family member 26B (220002_at), score: -0.7 KIR3DX1killer cell immunoglobulin-like receptor, three domains, X1 (216428_x_at), score: 0.6 KLHL3kelch-like 3 (Drosophila) (221221_s_at), score: -0.45 KMOkynurenine 3-monooxygenase (kynurenine 3-hydroxylase) (205307_s_at), score: 0.54 KRT8keratin 8 (209008_x_at), score: -0.52 KRT86keratin 86 (215189_at), score: -0.5 KRT8P12keratin 8 pseudogene 12 (222060_at), score: -0.86 KSR1kinase suppressor of ras 1 (213769_at), score: -0.66 LATS1LATS, large tumor suppressor, homolog 1 (Drosophila) (219813_at), score: 0.63 LILRA5leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 5 (215838_at), score: -0.54 LILRB3leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3 (211133_x_at), score: -0.42 LILRB4leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 4 (210152_at), score: -0.61 LINS1lines homolog 1 (Drosophila) (220121_at), score: 0.41 LOC100128919similar to HSPC157 (219865_at), score: 0.6 LOC100133503similar to LRRC37A3 protein (220219_s_at), score: -0.42 LOC100133748similar to GTF2IRD2 protein (215569_at), score: -0.64 LOC149478hypothetical protein LOC149478 (215462_at), score: -0.49 LOC149501similar to keratin 8 (216821_at), score: -0.52 LOC388152hypothetical LOC388152 (220602_s_at), score: -0.43 LOC80054hypothetical LOC80054 (220465_at), score: -0.4 LSRlipolysis stimulated lipoprotein receptor (208190_s_at), score: 0.52 LTBP4latent transforming growth factor beta binding protein 4 (210628_x_at), score: -0.59 MAPK8IP1mitogen-activated protein kinase 8 interacting protein 1 (213013_at), score: -0.66 MAPK8IP2mitogen-activated protein kinase 8 interacting protein 2 (205050_s_at), score: -0.56 MAPK8IP3mitogen-activated protein kinase 8 interacting protein 3 (213178_s_at), score: -0.54 MARK1MAP/microtubule affinity-regulating kinase 1 (221047_s_at), score: -0.43 MERTKc-mer proto-oncogene tyrosine kinase (211913_s_at), score: -0.61 MGC13053hypothetical MGC13053 (211775_x_at), score: 0.47 MICAL3microtubule associated monoxygenase, calponin and LIM domain containing 3 (212715_s_at), score: 0.44 MOCS1molybdenum cofactor synthesis 1 (211673_s_at), score: -0.53 MSTP9macrophage stimulating, pseudogene 9 (213382_at), score: -0.43 MUC5ACmucin 5AC, oligomeric mucus/gel-forming (217182_at), score: -0.69 MYL10myosin, light chain 10, regulatory (221659_s_at), score: -0.44 MYO15Amyosin XVA (220288_at), score: -0.56 MYO1Amyosin IA (211916_s_at), score: -0.48 NCAM2neural cell adhesion molecule 2 (205669_at), score: -0.48 NDRG3NDRG family member 3 (221082_s_at), score: 0.43 NDUFA2NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2, 8kDa (213550_s_at), score: 0.42 NFATC2IPnuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 interacting protein (217526_at), score: 0.42 NOVA2neuro-oncological ventral antigen 2 (206477_s_at), score: -0.59 NPEPL1aminopeptidase-like 1 (89476_r_at), score: -0.5 NSBP1nucleosomal binding protein 1 (221606_s_at), score: 0.46 NUDT13nudix (nucleoside diphosphate linked moiety X)-type motif 13 (214136_at), score: 0.43 NUPL2nucleoporin like 2 (204003_s_at), score: -0.39 NXPH4neurexophilin 4 (221967_at), score: -0.46 OGFOD12-oxoglutarate and iron-dependent oxygenase domain containing 1 (221090_s_at), score: -0.5 OGFRopioid growth factor receptor (211512_s_at), score: -0.46 OPRL1opiate receptor-like 1 (206564_at), score: -0.72 OR1E2olfactory receptor, family 1, subfamily E, member 2 (208587_s_at), score: -0.43 OR3A2olfactory receptor, family 3, subfamily A, member 2 (221386_at), score: -0.39 PARD6Apar-6 partitioning defective 6 homolog alpha (C. elegans) (205245_at), score: -0.42 PAX4paired box 4 (207867_at), score: 0.6 PCNXL2pecanex-like 2 (Drosophila) (39650_s_at), score: -0.56 PCOTHprostate collagen triple helix (222277_at), score: 0.6 PCSK5proprotein convertase subtilisin/kexin type 5 (205559_s_at), score: -0.4 PEX13peroxisomal biogenesis factor 13 (205246_at), score: 0.45 PGK2phosphoglycerate kinase 2 (217009_at), score: -0.62 PHC1polyhomeotic homolog 1 (Drosophila) (218338_at), score: 0.44 PHF2PHD finger protein 2 (212726_at), score: 0.57 PIK3R3phosphoinositide-3-kinase, regulatory subunit 3 (gamma) (202743_at), score: 0.51 PIN1Lpeptidylprolyl cis/trans isomerase, NIMA-interacting 1-like (pseudogene) (207582_at), score: -0.48 PIPOXpipecolic acid oxidase (221605_s_at), score: -0.55 PKLRpyruvate kinase, liver and RBC (222078_at), score: -0.68 PLCG2phospholipase C, gamma 2 (phosphatidylinositol-specific) (204613_at), score: -0.52 PLXNB3plexin B3 (205957_at), score: -0.54 PNMAL1PNMA-like 1 (218824_at), score: 0.69 POLR2J4polymerase (RNA) II (DNA directed) polypeptide J4, pseudogene (217610_at), score: -0.57 POM121L2POM121 membrane glycoprotein-like 2 (rat) (216582_at), score: -0.8 PRSS7protease, serine, 7 (enterokinase) (217269_s_at), score: -0.44 PTH1Rparathyroid hormone 1 receptor (205911_at), score: -0.47 PTPN2protein tyrosine phosphatase, non-receptor type 2 (213136_at), score: -0.4 RAP1GAPRAP1 GTPase activating protein (203911_at), score: -0.42 RBAKRB-associated KRAB zinc finger (219214_s_at), score: -0.43 RBM12BRNA binding motif protein 12B (51228_at), score: 0.65 RBM38RNA binding motif protein 38 (212430_at), score: -0.7 REG3Aregenerating islet-derived 3 alpha (205815_at), score: -0.48 RETret proto-oncogene (205879_x_at), score: 0.44 RLN1relaxin 1 (211753_s_at), score: -0.47 RNF121ring finger protein 121 (219021_at), score: -0.8 RNF146ring finger protein 146 (221430_s_at), score: -0.49 ROS1c-ros oncogene 1 , receptor tyrosine kinase (207569_at), score: -0.55 RPL10P16ribosomal protein L10 pseudogene 16 (217680_x_at), score: -0.45 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: -0.45 RPS6KA1ribosomal protein S6 kinase, 90kDa, polypeptide 1 (203379_at), score: -0.72 RPS6KA4ribosomal protein S6 kinase, 90kDa, polypeptide 4 (204632_at), score: 0.43 RPS6KA5ribosomal protein S6 kinase, 90kDa, polypeptide 5 (204633_s_at), score: 0.51 RSAD1radical S-adenosyl methionine domain containing 1 (218307_at), score: 0.41 SCLYselenocysteine lyase (221575_at), score: 0.93 SEC14L2SEC14-like 2 (S. cerevisiae) (204541_at), score: -0.4 SEPT5septin 5 (209767_s_at), score: -0.72 SERPINA1serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 (211429_s_at), score: 0.41 SERPINB13serpin peptidase inhibitor, clade B (ovalbumin), member 13 (211362_s_at), score: -0.39 SH2D3ASH2 domain containing 3A (222169_x_at), score: -0.49 SIGLEC1sialic acid binding Ig-like lectin 1, sialoadhesin (44673_at), score: -0.39 SIX6SIX homeobox 6 (207250_at), score: -0.61 SLC17A7solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 7 (204230_s_at), score: 0.44 SLC25A28solute carrier family 25, member 28 (221432_s_at), score: 0.46 SLC26A10solute carrier family 26, member 10 (214951_at), score: -0.54 SLC43A1solute carrier family 43, member 1 (204394_at), score: -0.41 SLC5A5solute carrier family 5 (sodium iodide symporter), member 5 (211123_at), score: -0.54 SMA4glucuronidase, beta pseudogene (215599_at), score: 0.41 SOCS3suppressor of cytokine signaling 3 (206359_at), score: -0.44 SORBS1sorbin and SH3 domain containing 1 (218087_s_at), score: -0.56 SP1Sp1 transcription factor (214732_at), score: 0.44 SRD5A3steroid 5 alpha-reductase 3 (218800_at), score: 0.51 ST3GAL4ST3 beta-galactoside alpha-2,3-sialyltransferase 4 (203759_at), score: -0.39 ST8SIA3ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 (208064_s_at), score: -0.71 STX17syntaxin 17 (218666_s_at), score: 0.57 SUV39H2suppressor of variegation 3-9 homolog 2 (Drosophila) (219262_at), score: 0.42 SYMPKsymplekin (202339_at), score: -0.41 SYNE2spectrin repeat containing, nuclear envelope 2 (202761_s_at), score: 0.52 SYT1synaptotagmin I (203999_at), score: 0.49 SYT5synaptotagmin V (206161_s_at), score: -0.48 TAF15TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa (202840_at), score: -0.58 TAL1T-cell acute lymphocytic leukemia 1 (206283_s_at), score: -0.49 TBX10T-box 10 (207689_at), score: -0.48 TEX264testis expressed 264 (218548_x_at), score: -0.44 TGM4transglutaminase 4 (prostate) (217566_s_at), score: -0.49 TICAM1toll-like receptor adaptor molecule 1 (213191_at), score: -0.43 TLE6transducin-like enhancer of split 6 (E(sp1) homolog, Drosophila) (222219_s_at), score: -0.49 TMEM132Atransmembrane protein 132A (218834_s_at), score: -0.41 TMEM30Btransmembrane protein 30B (213285_at), score: 0.44 TMEM80transmembrane protein 80 (221951_at), score: -0.42 TMPRSS6transmembrane protease, serine 6 (214955_at), score: -0.87 TMSB4Ythymosin beta 4, Y-linked (206769_at), score: -0.55 TNFRSF14tumor necrosis factor receptor superfamily, member 14 (herpesvirus entry mediator) (209354_at), score: -0.4 TOM1target of myb1 (chicken) (202807_s_at), score: -0.44 TPK1thiamin pyrophosphokinase 1 (221218_s_at), score: 0.56 TRAF4TNF receptor-associated factor 4 (202871_at), score: -0.43 TRAF6TNF receptor-associated factor 6 (205558_at), score: 0.44 TRIM62tripartite motif-containing 62 (219272_at), score: 0.49 TRMT61AtRNA methyltransferase 61 homolog A (S. cerevisiae) (52741_at), score: -0.46 TRMUtRNA 5-methylaminomethyl-2-thiouridylate methyltransferase (213634_s_at), score: -0.49 TRPC1transient receptor potential cation channel, subfamily C, member 1 (205802_at), score: 0.43 TRPV4transient receptor potential cation channel, subfamily V, member 4 (219516_at), score: -0.64 TSEN2tRNA splicing endonuclease 2 homolog (S. cerevisiae) (219581_at), score: 0.48 TSKStestis-specific serine kinase substrate (220545_s_at), score: -0.52 TSPAN9tetraspanin 9 (220968_s_at), score: -0.42 UTF1undifferentiated embryonic cell transcription factor 1 (208275_x_at), score: -0.48 VCPIP1valosin containing protein (p97)/p47 complex interacting protein 1 (219810_at), score: -0.59 WDR91WD repeat domain 91 (218971_s_at), score: -0.43 WFDC8WAP four-disulfide core domain 8 (215276_at), score: -0.47 XYLBxylulokinase homolog (H. influenzae) (214776_x_at), score: -0.52 YPEL1yippee-like 1 (Drosophila) (206063_x_at), score: -0.69 ZBTB10zinc finger and BTB domain containing 10 (219312_s_at), score: 0.64 ZBTB22zinc finger and BTB domain containing 22 (213081_at), score: -0.4 ZBTB3zinc finger and BTB domain containing 3 (220391_at), score: -0.46 ZDHHC11zinc finger, DHHC-type containing 11 (221646_s_at), score: -0.46 ZFYVE9zinc finger, FYVE domain containing 9 (204893_s_at), score: 0.44 ZNF192zinc finger protein 192 (206579_at), score: 0.66 ZNF20zinc finger protein 20 (213916_at), score: 0.46 ZNF202zinc finger protein 202 (204327_s_at), score: 0.42 ZNF214zinc finger protein 214 (220497_at), score: 0.86 ZNF236zinc finger protein 236 (47571_at), score: 0.52 ZNF510zinc finger protein 510 (206053_at), score: 0.43 ZNF571zinc finger protein 571 (206648_at), score: 0.53 ZNF839zinc finger protein 839 (219086_at), score: 0.43 ZSCAN16zinc finger and SCAN domain containing 16 (219676_at), score: 0.43 ZXDAzinc finger, X-linked, duplicated A (215263_at), score: 0.44
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485951.cel | 16 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486111.cel | 24 | 6 | Cycle | hgu133a2 | none | Cycle 1 |