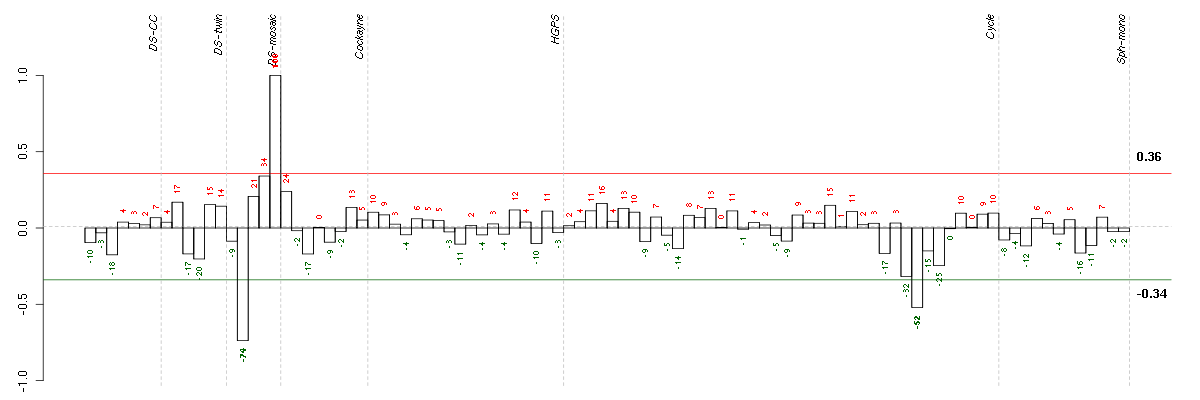

Under-expression is coded with green,
over-expression with red color.

immune system process
Any process involved in the development or functioning of the immune system, an organismal system for calibrated responses to potential internal or invasive threats.
defense response
Reactions, triggered in response to the presence of a foreign body or the occurrence of an injury, which result in restriction of damage to the organism attacked or prevention/recovery from the infection caused by the attack.
cell adhesion
The attachment of a cell, either to another cell or to an underlying substrate such as the extracellular matrix, via cell adhesion molecules.
response to stress
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a disturbance in organismal or cellular homeostasis, usually, but not necessarily, exogenous (e.g. temperature, humidity, ionizing radiation).
immune response
Any immune system process that functions in the calibrated response of an organism to a potential internal or invasive threat.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
biological adhesion
The attachment of a cell or organism to a substrate or other organism.
innate immune response
Innate immune responses are defense responses mediated by germline encoded components that directly recognize components of potential pathogens.
response to stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
all
This term is the most general term possible
cell adhesion
The attachment of a cell, either to another cell or to an underlying substrate such as the extracellular matrix, via cell adhesion molecules.
immune response
Any immune system process that functions in the calibrated response of an organism to a potential internal or invasive threat.
innate immune response
Innate immune responses are defense responses mediated by germline encoded components that directly recognize components of potential pathogens.

extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
all
This term is the most general term possible
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.

ABOABO blood group (transferase A, alpha 1-3-N-acetylgalactosaminyltransferase; transferase B, alpha 1-3-galactosyltransferase) (216716_at), score: 0.48 ADAMTS12ADAM metallopeptidase with thrombospondin type 1 motif, 12 (221421_s_at), score: 0.87 ADH1Balcohol dehydrogenase 1B (class I), beta polypeptide (209612_s_at), score: -0.54 AFF4AF4/FMR2 family, member 4 (219199_at), score: 0.47 AGRNagrin (212285_s_at), score: -0.5 AGTangiotensinogen (serpin peptidase inhibitor, clade A, member 8) (202834_at), score: -0.49 ALDOBaldolase B, fructose-bisphosphate (214423_x_at), score: 0.51 ALDOCaldolase C, fructose-bisphosphate (202022_at), score: -0.48 ANGPTL2angiopoietin-like 2 (213004_at), score: -0.71 AP1S1adaptor-related protein complex 1, sigma 1 subunit (205195_at), score: 0.47 APOL1apolipoprotein L, 1 (209546_s_at), score: -0.56 APOL3apolipoprotein L, 3 (221087_s_at), score: -0.6 APOL6apolipoprotein L, 6 (219716_at), score: -0.73 ARFRP1ADP-ribosylation factor related protein 1 (215984_s_at), score: 0.58 ARHGAP26Rho GTPase activating protein 26 (205068_s_at), score: -0.52 ARHGDIARho GDP dissociation inhibitor (GDI) alpha (213606_s_at), score: 0.47 ARPC4actin related protein 2/3 complex, subunit 4, 20kDa (211672_s_at), score: 0.49 ASAP3ArfGAP with SH3 domain, ankyrin repeat and PH domain 3 (219103_at), score: -0.61 B3GNT2UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2 (219326_s_at), score: 0.51 B9D1B9 protein domain 1 (210534_s_at), score: -0.71 BAT2D1BAT2 domain containing 1 (211947_s_at), score: 0.56 BAZ1Abromodomain adjacent to zinc finger domain, 1A (217985_s_at), score: 0.47 BCAMbasal cell adhesion molecule (Lutheran blood group) (40093_at), score: -0.68 BCHEbutyrylcholinesterase (205433_at), score: 0.67 BTRCbeta-transducin repeat containing (216091_s_at), score: 0.58 C11orf41chromosome 11 open reading frame 41 (214772_at), score: 0.62 C14orf159chromosome 14 open reading frame 159 (218298_s_at), score: -0.49 C1Rcomplement component 1, r subcomponent (212067_s_at), score: -0.55 CA11carbonic anhydrase XI (209726_at), score: -0.91 CACNA2D1calcium channel, voltage-dependent, alpha 2/delta subunit 1 (207050_at), score: 0.47 CASP1caspase 1, apoptosis-related cysteine peptidase (interleukin 1, beta, convertase) (211368_s_at), score: -0.9 CCL11chemokine (C-C motif) ligand 11 (210133_at), score: -0.53 CCND1cyclin D1 (208711_s_at), score: 0.54 CD302CD302 molecule (203799_at), score: -0.61 CD44CD44 molecule (Indian blood group) (210916_s_at), score: 0.53 CEBPBCCAAT/enhancer binding protein (C/EBP), beta (212501_at), score: -0.53 CFIcomplement factor I (203854_at), score: -0.9 CHMLchoroideremia-like (Rab escort protein 2) (206079_at), score: 0.49 CHST11carbohydrate (chondroitin 4) sulfotransferase 11 (219634_at), score: 0.58 CHST5carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 5 (219182_at), score: -0.51 CMAHcytidine monophosphate-N-acetylneuraminic acid hydroxylase (CMP-N-acetylneuraminate monooxygenase) pseudogene (205518_s_at), score: 0.53 COL14A1collagen, type XIV, alpha 1 (212865_s_at), score: -0.78 COL21A1collagen, type XXI, alpha 1 (208096_s_at), score: -0.63 CPA3carboxypeptidase A3 (mast cell) (205624_at), score: -0.55 CRKRSCdc2-related kinase, arginine/serine-rich (219226_at), score: 0.54 CRLF1cytokine receptor-like factor 1 (206315_at), score: 0.47 CSPG4chondroitin sulfate proteoglycan 4 (214297_at), score: 0.48 CTDSP2CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 (203445_s_at), score: -0.56 CTSHcathepsin H (202295_s_at), score: -0.52 CTSOcathepsin O (203758_at), score: -0.48 CUGBP1CUG triplet repeat, RNA binding protein 1 (204113_at), score: 0.55 CYTH3cytohesin 3 (206523_at), score: 0.58 CYTIPcytohesin 1 interacting protein (209606_at), score: 0.9 DAPK3death-associated protein kinase 3 (203890_s_at), score: 0.55 DCLK1doublecortin-like kinase 1 (205399_at), score: -0.52 DENND2ADENN/MADD domain containing 2A (53991_at), score: -0.54 DENND3DENN/MADD domain containing 3 (212974_at), score: 0.51 DHX9DEAH (Asp-Glu-Ala-His) box polypeptide 9 (212105_s_at), score: 0.58 DKFZp434H1419hypothetical protein DKFZp434H1419 (214717_at), score: 0.58 DKFZP586H2123regeneration associated muscle protease (213661_at), score: -0.57 DOM3Zdom-3 homolog Z (C. elegans) (215982_s_at), score: -0.65 DOPEY2dopey family member 2 (205248_at), score: 0.77 DSC2desmocollin 2 (204751_x_at), score: 0.56 ECHDC2enoyl Coenzyme A hydratase domain containing 2 (218552_at), score: -0.72 ECSITECSIT homolog (Drosophila) (218225_at), score: -0.62 EDN1endothelin 1 (218995_s_at), score: 0.55 EFHC1EF-hand domain (C-terminal) containing 1 (219833_s_at), score: -0.5 EGFepidermal growth factor (beta-urogastrone) (206254_at), score: -0.58 EIF2S1eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa (201143_s_at), score: 0.48 ELOVL2elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 2 (213712_at), score: -0.77 ELTD1EGF, latrophilin and seven transmembrane domain containing 1 (219134_at), score: 0.46 ERAP1endoplasmic reticulum aminopeptidase 1 (214012_at), score: -0.66 ERN1endoplasmic reticulum to nucleus signaling 1 (207061_at), score: 0.6 ETS1v-ets erythroblastosis virus E26 oncogene homolog 1 (avian) (214447_at), score: 0.47 EZRezrin (208621_s_at), score: 0.61 FAM102Afamily with sequence similarity 102, member A (212400_at), score: -0.51 FAM155Afamily with sequence similarity 155, member A (214825_at), score: 0.51 FEZ1fasciculation and elongation protein zeta 1 (zygin I) (203562_at), score: -0.55 FN1fibronectin 1 (214701_s_at), score: 0.48 FNBP1formin binding protein 1 (212288_at), score: -0.6 FOSv-fos FBJ murine osteosarcoma viral oncogene homolog (209189_at), score: -0.97 GBP2guanylate binding protein 2, interferon-inducible (202748_at), score: -0.55 GCH1GTP cyclohydrolase 1 (204224_s_at), score: -0.73 GDF15growth differentiation factor 15 (221577_x_at), score: -0.55 GGT1gamma-glutamyltransferase 1 (209919_x_at), score: -0.54 GOLGA8Ggolgi autoantigen, golgin subfamily a, 8G (222149_x_at), score: -0.5 GOLSYNGolgi-localized protein (218692_at), score: -0.49 GP1BBglycoprotein Ib (platelet), beta polypeptide (206655_s_at), score: -0.58 GPC4glypican 4 (204983_s_at), score: -0.67 GPR37G protein-coupled receptor 37 (endothelin receptor type B-like) (209631_s_at), score: -0.74 GTF2A1Lgeneral transcription factor IIA, 1-like (213413_at), score: -0.51 GTF2Igeneral transcription factor II, i (210892_s_at), score: 0.57 HAPLN1hyaluronan and proteoglycan link protein 1 (205523_at), score: 0.63 HHEXhematopoietically expressed homeobox (215933_s_at), score: -0.87 HIPK2homeodomain interacting protein kinase 2 (219028_at), score: 0.48 HIST3H2Ahistone cluster 3, H2a (221582_at), score: -0.62 HMOX1heme oxygenase (decycling) 1 (203665_at), score: -0.49 HN1Lhematological and neurological expressed 1-like (212109_at), score: 0.46 HOXB9homeobox B9 (216417_x_at), score: 0.99 HS1BP3HCLS1 binding protein 3 (219020_at), score: -0.7 IFI35interferon-induced protein 35 (209417_s_at), score: -0.49 IL15RAinterleukin 15 receptor, alpha (207375_s_at), score: -0.55 IL1R1interleukin 1 receptor, type I (202948_at), score: -0.52 IL4Rinterleukin 4 receptor (203233_at), score: 0.47 INAinternexin neuronal intermediate filament protein, alpha (204465_s_at), score: 0.49 INHBEinhibin, beta E (210587_at), score: -0.81 INPP5Binositol polyphosphate-5-phosphatase, 75kDa (213804_at), score: -0.51 IRF1interferon regulatory factor 1 (202531_at), score: -0.66 ISYNA1inositol-3-phosphate synthase 1 (222240_s_at), score: -0.48 ITGA6integrin, alpha 6 (215177_s_at), score: 0.53 ITPKCinositol 1,4,5-trisphosphate 3-kinase C (213076_at), score: -0.53 KCNN2potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2 (220116_at), score: 0.7 KCNN4potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 (204401_at), score: 0.45 KCTD20potassium channel tetramerisation domain containing 20 (214849_at), score: 0.5 KIAA0556KIAA0556 (213185_at), score: -0.62 KIAA0644KIAA0644 gene product (205150_s_at), score: -0.76 L1CAML1 cell adhesion molecule (204584_at), score: -0.62 LAMA5laminin, alpha 5 (210150_s_at), score: -0.6 LATS1LATS, large tumor suppressor, homolog 1 (Drosophila) (219813_at), score: 0.52 LETMD1LETM1 domain containing 1 (207170_s_at), score: -0.5 LHPPphospholysine phosphohistidine inorganic pyrophosphate phosphatase (218523_at), score: -0.71 LILRA5leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 5 (215838_at), score: 0.54 LOC100128414similar to fibrinogen silencer binding protein (220549_at), score: 0.51 LOC100133105hypothetical protein LOC100133105 (214237_x_at), score: 0.48 LOC730092RRN3 RNA polymerase I transcription factor homolog (S. cerevisiae) pseudogene (216908_x_at), score: 0.61 LRRN3leucine rich repeat neuronal 3 (209841_s_at), score: 0.55 LZTFL1leucine zipper transcription factor-like 1 (218437_s_at), score: -0.63 MACROD1MACRO domain containing 1 (219188_s_at), score: -0.75 MAN1C1mannosidase, alpha, class 1C, member 1 (218918_at), score: -0.78 MAP2K5mitogen-activated protein kinase kinase 5 (211370_s_at), score: -0.69 MAXMYC associated factor X (210734_x_at), score: 0.5 MAZMYC-associated zinc finger protein (purine-binding transcription factor) (207824_s_at), score: 0.57 MCAMmelanoma cell adhesion molecule (210869_s_at), score: 0.51 ME3malic enzyme 3, NADP(+)-dependent, mitochondrial (204663_at), score: -0.73 MED31mediator complex subunit 31 (219318_x_at), score: 0.52 MEGF9multiple EGF-like-domains 9 (212830_at), score: -0.65 MFAP4microfibrillar-associated protein 4 (212713_at), score: -0.84 MFN2mitofusin 2 (216205_s_at), score: 0.61 MFSD6major facilitator superfamily domain containing 6 (219858_s_at), score: -0.62 MGMTO-6-methylguanine-DNA methyltransferase (204880_at), score: -0.48 MMP14matrix metallopeptidase 14 (membrane-inserted) (202827_s_at), score: 0.5 MOCOSmolybdenum cofactor sulfurase (219959_at), score: -0.66 MPDU1mannose-P-dolichol utilization defect 1 (209208_at), score: 0.47 MRASmuscle RAS oncogene homolog (206538_at), score: 0.55 MRPS6mitochondrial ribosomal protein S6 (213167_s_at), score: 0.61 MSR1macrophage scavenger receptor 1 (208423_s_at), score: 0.46 MTMR12myotubularin related protein 12 (220953_s_at), score: 0.46 MVKmevalonate kinase (36907_at), score: -0.51 MX1myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) (202086_at), score: -0.54 MYO1Dmyosin ID (212338_at), score: -0.49 NAB2NGFI-A binding protein 2 (EGR1 binding protein 2) (216017_s_at), score: 0.46 NAGLUN-acetylglucosaminidase, alpha- (204360_s_at), score: -0.54 NCOA1nuclear receptor coactivator 1 (209106_at), score: -0.57 NEDD4Lneural precursor cell expressed, developmentally down-regulated 4-like (212445_s_at), score: 0.55 NET1neuroepithelial cell transforming 1 (201830_s_at), score: 0.51 NF2neurofibromin 2 (merlin) (211017_s_at), score: 0.51 NHEJ1nonhomologous end-joining factor 1 (219418_at), score: -0.52 NINJ1ninjurin 1 (203045_at), score: -0.54 NOS1nitric oxide synthase 1 (neuronal) (207309_at), score: -0.51 NPTX1neuronal pentraxin I (204684_at), score: 0.52 NR1H3nuclear receptor subfamily 1, group H, member 3 (203920_at), score: -0.72 NRIP3nuclear receptor interacting protein 3 (219557_s_at), score: 0.46 NUMA1nuclear mitotic apparatus protein 1 (214251_s_at), score: 0.5 OAS12',5'-oligoadenylate synthetase 1, 40/46kDa (205552_s_at), score: -0.5 OLFML2Aolfactomedin-like 2A (213075_at), score: -0.81 OLR1oxidized low density lipoprotein (lectin-like) receptor 1 (210004_at), score: 0.68 OSGEPO-sialoglycoprotein endopeptidase (209450_at), score: 0.48 OTUD3OTU domain containing 3 (213216_at), score: 0.55 PAK1p21 protein (Cdc42/Rac)-activated kinase 1 (209615_s_at), score: 0.47 PAPPA2pappalysin 2 (213332_at), score: -0.82 PARP12poly (ADP-ribose) polymerase family, member 12 (218543_s_at), score: -0.57 PCK2phosphoenolpyruvate carboxykinase 2 (mitochondrial) (202847_at), score: -0.5 PDE1Cphosphodiesterase 1C, calmodulin-dependent 70kDa (207303_at), score: 0.73 PDGFRLplatelet-derived growth factor receptor-like (205226_at), score: -0.55 PDK2pyruvate dehydrogenase kinase, isozyme 2 (202590_s_at), score: -0.52 PIGZphosphatidylinositol glycan anchor biosynthesis, class Z (220041_at), score: -0.55 PIP5K1Aphosphatidylinositol-4-phosphate 5-kinase, type I, alpha (211205_x_at), score: 0.52 PKP2plakophilin 2 (207717_s_at), score: -0.67 PLA2G4Cphospholipase A2, group IVC (cytosolic, calcium-independent) (209785_s_at), score: -0.49 PLEKHB2pleckstrin homology domain containing, family B (evectins) member 2 (201411_s_at), score: 0.49 PLEKHM1pleckstrin homology domain containing, family M (with RUN domain) member 1 (212717_at), score: -0.5 PLS1plastin 1 (I isoform) (205190_at), score: 0.59 PLTPphospholipid transfer protein (202075_s_at), score: -0.64 PNRC1proline-rich nuclear receptor coactivator 1 (209034_at), score: -0.53 PPARGperoxisome proliferator-activated receptor gamma (208510_s_at), score: -0.5 PPP2R5Dprotein phosphatase 2, regulatory subunit B', delta isoform (211159_s_at), score: 0.58 PRKG1protein kinase, cGMP-dependent, type I (207119_at), score: -0.48 PSG3pregnancy specific beta-1-glycoprotein 3 (215821_x_at), score: 0.47 PSG9pregnancy specific beta-1-glycoprotein 9 (209594_x_at), score: 0.48 PSMB10proteasome (prosome, macropain) subunit, beta type, 10 (202659_at), score: -0.58 PSME4proteasome (prosome, macropain) activator subunit 4 (212220_at), score: 0.67 PTBP1polypyrimidine tract binding protein 1 (212016_s_at), score: 0.47 PTGDSprostaglandin D2 synthase 21kDa (brain) (212187_x_at), score: -0.87 PTGER4prostaglandin E receptor 4 (subtype EP4) (204897_at), score: -0.63 PTPN2protein tyrosine phosphatase, non-receptor type 2 (213136_at), score: -0.56 PTPN21protein tyrosine phosphatase, non-receptor type 21 (205438_at), score: 0.46 PTPRAprotein tyrosine phosphatase, receptor type, A (213799_s_at), score: 0.58 PTPREprotein tyrosine phosphatase, receptor type, E (221840_at), score: -0.62 PTRFpolymerase I and transcript release factor (208790_s_at), score: 0.51 PXNpaxillin (211823_s_at), score: 0.47 RANGAP1Ran GTPase activating protein 1 (212125_at), score: 0.55 RARRES3retinoic acid receptor responder (tazarotene induced) 3 (204070_at), score: -1 RASA3RAS p21 protein activator 3 (206220_s_at), score: 0.63 RGS17regulator of G-protein signaling 17 (220334_at), score: -0.52 ROBO3roundabout, axon guidance receptor, homolog 3 (Drosophila) (219550_at), score: -0.51 ROR2receptor tyrosine kinase-like orphan receptor 2 (205578_at), score: -0.65 SCARB1scavenger receptor class B, member 1 (201819_at), score: -0.76 SCG2secretogranin II (chromogranin C) (204035_at), score: -0.57 SCRG1scrapie responsive protein 1 (205475_at), score: -0.62 SEC14L1SEC14-like 1 (S. cerevisiae) (202082_s_at), score: 0.58 SELPLGselectin P ligand (209879_at), score: 0.67 SERPINB9serpin peptidase inhibitor, clade B (ovalbumin), member 9 (209723_at), score: -0.52 SGCGsarcoglycan, gamma (35kDa dystrophin-associated glycoprotein) (207302_at), score: -0.58 SH2D3ASH2 domain containing 3A (222169_x_at), score: 0.51 SIX2SIX homeobox 2 (206510_at), score: -0.57 SLC16A3solute carrier family 16, member 3 (monocarboxylic acid transporter 4) (202855_s_at), score: 0.48 SLC26A6solute carrier family 26, member 6 (221572_s_at), score: -0.71 SLC27A3solute carrier family 27 (fatty acid transporter), member 3 (222217_s_at), score: -0.64 SLC29A1solute carrier family 29 (nucleoside transporters), member 1 (201801_s_at), score: 0.73 SLCO2A1solute carrier organic anion transporter family, member 2A1 (204368_at), score: -0.51 SOD3superoxide dismutase 3, extracellular (205236_x_at), score: -0.64 SONSON DNA binding protein (201085_s_at), score: 0.46 SPATA7spermatogenesis associated 7 (219583_s_at), score: -0.5 SPIN1spindlin 1 (217813_s_at), score: 0.47 SPSB3splA/ryanodine receptor domain and SOCS box containing 3 (46256_at), score: -0.53 STAT2signal transducer and activator of transcription 2, 113kDa (205170_at), score: 0.78 SULF1sulfatase 1 (212353_at), score: -0.54 SULT1A3sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 (210580_x_at), score: -0.81 SVEP1sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 (213247_at), score: -0.63 TANC2tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 (208425_s_at), score: 0.77 TAPBPLTAP binding protein-like (218747_s_at), score: -0.74 TBC1D12TBC1 domain family, member 12 (221858_at), score: -0.6 TBX10T-box 10 (207689_at), score: 0.49 TGFBR1transforming growth factor, beta receptor 1 (206943_at), score: 0.61 THRAthyroid hormone receptor, alpha (erythroblastic leukemia viral (v-erb-a) oncogene homolog, avian) (35846_at), score: -0.51 TLR3toll-like receptor 3 (206271_at), score: -0.65 TM6SF1transmembrane 6 superfamily member 1 (219892_at), score: -0.55 TM7SF2transmembrane 7 superfamily member 2 (210130_s_at), score: -0.74 TMEM132Atransmembrane protein 132A (218834_s_at), score: -0.5 TMEM160transmembrane protein 160 (219219_at), score: 0.48 TMEM48transmembrane protein 48 (218073_s_at), score: 0.62 TNFAIP6tumor necrosis factor, alpha-induced protein 6 (206026_s_at), score: -0.54 TNFRSF14tumor necrosis factor receptor superfamily, member 14 (herpesvirus entry mediator) (209354_at), score: -0.58 TNFSF9tumor necrosis factor (ligand) superfamily, member 9 (206907_at), score: -0.65 TOM1target of myb1 (chicken) (202807_s_at), score: -0.67 TOM1L1target of myb1 (chicken)-like 1 (204485_s_at), score: -0.51 TOM1L2target of myb1-like 2 (chicken) (214840_at), score: 0.76 TOP3Atopoisomerase (DNA) III alpha (214300_s_at), score: 0.65 TPCN1two pore segment channel 1 (217914_at), score: -0.5 TRIM2tripartite motif-containing 2 (202342_s_at), score: -0.52 TRIM3tripartite motif-containing 3 (204911_s_at), score: 0.52 UPF1UPF1 regulator of nonsense transcripts homolog (yeast) (211168_s_at), score: 0.51 USP21ubiquitin specific peptidase 21 (218367_x_at), score: -0.52 USP36ubiquitin specific peptidase 36 (220370_s_at), score: 0.47 VAV2vav 2 guanine nucleotide exchange factor (205536_at), score: 0.49 VCAM1vascular cell adhesion molecule 1 (203868_s_at), score: -0.69 WDR19WD repeat domain 19 (220917_s_at), score: -0.56 XYLT1xylosyltransferase I (213725_x_at), score: 0.49 ZNF467zinc finger protein 467 (214746_s_at), score: -0.61 ZNF688zinc finger protein 688 (213527_s_at), score: -0.66 ZNF711zinc finger protein 711 (207781_s_at), score: -0.61 ZRSR2zinc finger (CCCH type), RNA-binding motif and serine/arginine rich 2 (213876_x_at), score: -0.63
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| 46B.CEL | 2 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 2 |
| E-TABM-263-raw-cel-1515486291.cel | 33 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 47C.CEL | 5 | 3 | DS-mosaic | hgu133plus2 | Down mosaic | DS-mosaic 5 |