
Under-expression is coded with green,
over-expression with red color.


plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
cytoplasm
All of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
secretory granule
A small subcellular vesicle, surrounded by a membrane, that is formed from the Golgi apparatus and contains a highly concentrated protein destined for secretion. Secretory granules move towards the periphery of the cell, their membranes fuse with the cell membrane, and their protein load is exteriorized. Processing of the contained protein may take place in secretory granules.
cytoplasmic membrane-bounded vesicle
A membrane-bounded vesicle found in the cytoplasm of the cell.
platelet alpha granule
A secretory organelle found in blood platelets, which is unique in that it exhibits further compartmentalization and acquires its protein content via two distinct mechanisms: (1) biosynthesis predominantly at the megakaryocyte (MK) level (with some vestigial platelet synthesis) (e.g. platelet factor 4) and (2) endocytosis and pinocytosis at both the MK and circulating platelet levels (e.g. fibrinogen (Fg) and IgG).
cytoplasmic vesicle
A vesicle formed of membrane or protein, found in the cytoplasm of a cell.
vesicle
Any small, fluid-filled, spherical organelle enclosed by membrane or protein.
membrane-bounded vesicle
Any small, fluid-filled, spherical organelle enclosed by a lipid bilayer.
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
cytoplasmic vesicle
A vesicle formed of membrane or protein, found in the cytoplasm of a cell.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
cytoplasmic membrane-bounded vesicle
A membrane-bounded vesicle found in the cytoplasm of the cell.
cytoplasmic membrane-bounded vesicle
A membrane-bounded vesicle found in the cytoplasm of the cell.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
cytoplasmic vesicle
A vesicle formed of membrane or protein, found in the cytoplasm of a cell.

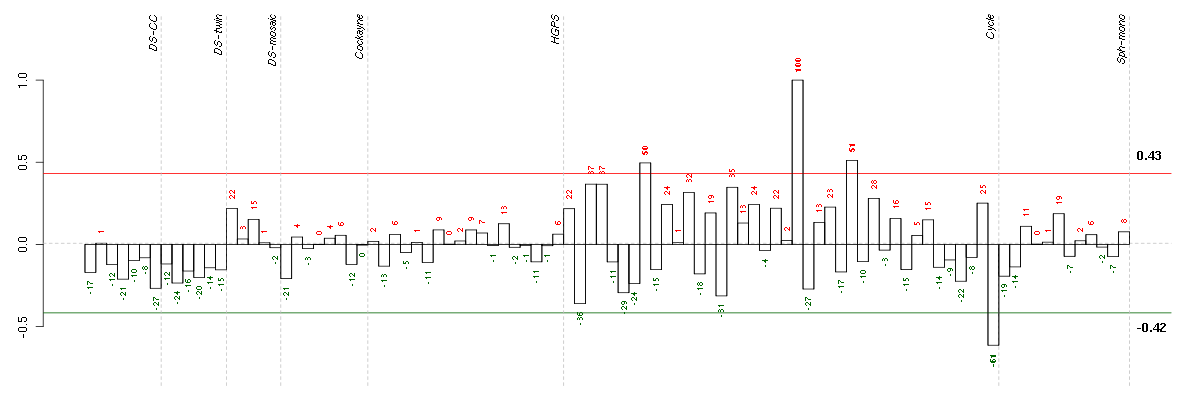
ABCA7ATP-binding cassette, sub-family A (ABC1), member 7 (219577_s_at), score: -0.53 ABLIM1actin binding LIM protein 1 (200965_s_at), score: 0.48 ABOABO blood group (transferase A, alpha 1-3-N-acetylgalactosaminyltransferase; transferase B, alpha 1-3-galactosyltransferase) (216716_at), score: -0.5 ABTB2ankyrin repeat and BTB (POZ) domain containing 2 (213497_at), score: -0.46 ACAD10acyl-Coenzyme A dehydrogenase family, member 10 (219986_s_at), score: -0.47 ADAM21ADAM metallopeptidase domain 21 (207665_at), score: 0.48 ADAMTS12ADAM metallopeptidase with thrombospondin type 1 motif, 12 (221421_s_at), score: -0.69 ADAMTS7ADAM metallopeptidase with thrombospondin type 1 motif, 7 (220705_s_at), score: -0.55 ADCK2aarF domain containing kinase 2 (221893_s_at), score: 0.44 ALDH3B1aldehyde dehydrogenase 3 family, member B1 (211004_s_at), score: -0.46 AMPHamphiphysin (205257_s_at), score: -0.48 ANK1ankyrin 1, erythrocytic (208353_x_at), score: -0.57 AP1G2adaptor-related protein complex 1, gamma 2 subunit (201613_s_at), score: -0.56 APOC4apolipoprotein C-IV (206738_at), score: -0.6 ARL17P1ADP-ribosylation factor-like 17 pseudogene 1 (210718_s_at), score: 0.45 ATHL1ATH1, acid trehalase-like 1 (yeast) (219359_at), score: -0.46 ATXN1ataxin 1 (203232_s_at), score: -0.47 BAIAP3BAI1-associated protein 3 (216356_x_at), score: -0.54 BANK1B-cell scaffold protein with ankyrin repeats 1 (219667_s_at), score: 0.44 BAT2HLA-B associated transcript 2 (212081_x_at), score: 0.47 BCANbrevican (91920_at), score: -0.49 BTNL3butyrophilin-like 3 (217207_s_at), score: -0.59 C10orf81chromosome 10 open reading frame 81 (219857_at), score: 0.93 C16orf57chromosome 16 open reading frame 57 (218060_s_at), score: 0.53 C19orf40chromosome 19 open reading frame 40 (214816_x_at), score: -0.46 C19orf61chromosome 19 open reading frame 61 (221335_x_at), score: 0.51 C1orf69chromosome 1 open reading frame 69 (215490_at), score: -0.6 C21orf2chromosome 21 open reading frame 2 (203996_s_at), score: -0.61 C3AR1complement component 3a receptor 1 (209906_at), score: -0.57 C6orf155chromosome 6 open reading frame 155 (220324_at), score: 0.5 C6orf162chromosome 6 open reading frame 162 (213314_at), score: -0.52 C7orf58chromosome 7 open reading frame 58 (220032_at), score: 0.45 CABC1chaperone, ABC1 activity of bc1 complex homolog (S. pombe) (218168_s_at), score: -0.58 CALB2calbindin 2 (205428_s_at), score: -0.46 CARD8caspase recruitment domain family, member 8 (204950_at), score: -0.45 CCDC101coiled-coil domain containing 101 (48117_at), score: -0.54 CCDC41coiled-coil domain containing 41 (219644_at), score: -0.53 CCDC49coiled-coil domain containing 49 (218655_s_at), score: 0.49 CD53CD53 molecule (203416_at), score: -0.49 CD79BCD79b molecule, immunoglobulin-associated beta (205297_s_at), score: -0.75 CDC14BCDC14 cell division cycle 14 homolog B (S. cerevisiae) (208022_s_at), score: -0.55 CDH4cadherin 4, type 1, R-cadherin (retinal) (220227_at), score: -0.49 CDKL3cyclin-dependent kinase-like 3 (219831_at), score: -0.48 CHN2chimerin (chimaerin) 2 (207486_x_at), score: -0.63 CHRNA10cholinergic receptor, nicotinic, alpha 10 (220210_at), score: -0.47 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: 0.51 COL11A2collagen, type XI, alpha 2 (216993_s_at), score: -0.54 CORO2Bcoronin, actin binding protein, 2B (209789_at), score: 0.51 CRKRSCdc2-related kinase, arginine/serine-rich (219226_at), score: 0.46 CTSEcathepsin E (205927_s_at), score: -0.59 CYP11B1cytochrome P450, family 11, subfamily B, polypeptide 1 (214610_at), score: -0.48 CYP3A4cytochrome P450, family 3, subfamily A, polypeptide 4 (205998_x_at), score: -0.46 DDX28DEAD (Asp-Glu-Ala-Asp) box polypeptide 28 (203785_s_at), score: 0.46 DNAJC4DnaJ (Hsp40) homolog, subfamily C, member 4 (206782_s_at), score: 0.64 DOPEY1dopey family member 1 (40612_at), score: -0.5 DSPPdentin sialophosphoprotein (221681_s_at), score: -0.54 EDARectodysplasin A receptor (220048_at), score: -0.67 EGFepidermal growth factor (beta-urogastrone) (206254_at), score: -0.47 ELA3Aelastase 3A, pancreatic (211738_x_at), score: -0.48 ELLelongation factor RNA polymerase II (204095_s_at), score: -0.46 ENO3enolase 3 (beta, muscle) (204483_at), score: -0.48 ENOX2ecto-NOX disulfide-thiol exchanger 2 (32042_at), score: -0.53 EPB41L5erythrocyte membrane protein band 4.1 like 5 (220977_x_at), score: -0.57 EPHA4EPH receptor A4 (206114_at), score: -0.6 EPHB4EPH receptor B4 (216680_s_at), score: -0.52 ERMAPerythroblast membrane-associated protein (Scianna blood group) (219905_at), score: -0.54 ETV7ets variant 7 (221680_s_at), score: -0.52 EVLEnah/Vasp-like (217838_s_at), score: 0.44 F7coagulation factor VII (serum prothrombin conversion accelerator) (207300_s_at), score: -0.52 FAM70Afamily with sequence similarity 70, member A (219895_at), score: 0.49 FBRSfibrosin (218255_s_at), score: -0.6 FBXL6F-box and leucine-rich repeat protein 6 (219189_at), score: 0.47 FBXO2F-box protein 2 (219305_x_at), score: -0.58 FCF1FCF1 small subunit (SSU) processome component homolog (S. cerevisiae) (219927_at), score: -0.45 FGL1fibrinogen-like 1 (205305_at), score: -0.58 FKBP10FK506 binding protein 10, 65 kDa (219249_s_at), score: -0.53 FLJ40113golgi autoantigen, golgin subfamily a-like pseudogene (213212_x_at), score: -0.51 FOXK2forkhead box K2 (203064_s_at), score: 0.53 GFOD2glucose-fructose oxidoreductase domain containing 2 (221028_s_at), score: 0.58 GGA3golgi associated, gamma adaptin ear containing, ARF binding protein 3 (209411_s_at), score: 0.46 GNL3LPguanine nucleotide binding protein-like 3 (nucleolar)-like pseudogene (220716_at), score: -0.51 GP1BAglycoprotein Ib (platelet), alpha polypeptide (207389_at), score: -0.59 GP6glycoprotein VI (platelet) (220336_s_at), score: 0.93 GPR144G protein-coupled receptor 144 (216289_at), score: -0.62 GRB10growth factor receptor-bound protein 10 (210999_s_at), score: -0.5 GRHL2grainyhead-like 2 (Drosophila) (219388_at), score: -0.46 HAB1B1 for mucin (215778_x_at), score: -0.53 HAMPhepcidin antimicrobial peptide (220491_at), score: -0.65 HBBhemoglobin, beta (211696_x_at), score: -0.6 HCRP1hepatocellular carcinoma-related HCRP1 (216176_at), score: -0.57 HDAC9histone deacetylase 9 (205659_at), score: 0.55 HECW1HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1 (215584_at), score: -0.63 HGFhepatocyte growth factor (hepapoietin A; scatter factor) (209960_at), score: 0.52 HIST1H4Dhistone cluster 1, H4d (208076_at), score: -0.5 HLA-DQB1major histocompatibility complex, class II, DQ beta 1 (211654_x_at), score: -0.55 HLA-DRB4major histocompatibility complex, class II, DR beta 4 (208306_x_at), score: -0.51 HNRNPH2heterogeneous nuclear ribonucleoprotein H2 (H') (201132_at), score: -0.45 HOXB5homeobox B5 (205601_s_at), score: -0.56 HPS4Hermansky-Pudlak syndrome 4 (54037_at), score: -0.52 HUNKhormonally up-regulated Neu-associated kinase (219535_at), score: -0.46 IDUAiduronidase, alpha-L- (205059_s_at), score: 0.52 IFNGinterferon, gamma (210354_at), score: -0.46 IGHG1immunoglobulin heavy constant gamma 1 (G1m marker) (211693_at), score: -0.48 INPP1inositol polyphosphate-1-phosphatase (202794_at), score: 0.49 IRF2interferon regulatory factor 2 (203275_at), score: 0.47 ISOC2isochorismatase domain containing 2 (218893_at), score: -0.54 ITGALintegrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide) (213475_s_at), score: -0.47 KAL1Kallmann syndrome 1 sequence (205206_at), score: -0.46 KCNJ14potassium inwardly-rectifying channel, subfamily J, member 14 (220776_at), score: -0.51 KCNMB4potassium large conductance calcium-activated channel, subfamily M, beta member 4 (219287_at), score: -0.52 KIAA0240KIAA0240 (38892_at), score: -0.47 KIAA0894KIAA0894 protein (207436_x_at), score: -0.51 KIAA1009KIAA1009 (206005_s_at), score: -0.5 KRT14keratin 14 (209351_at), score: 0.49 KRT86keratin 86 (215189_at), score: -0.72 LOC100133105hypothetical protein LOC100133105 (214237_x_at), score: -0.81 LOC100133748similar to GTF2IRD2 protein (215569_at), score: -0.58 LOC128192similar to peptidyl-Pro cis trans isomerase (217346_at), score: -0.51 LOC149478hypothetical protein LOC149478 (215462_at), score: -0.46 LOC149501similar to keratin 8 (216821_at), score: -0.46 LOC390940similar to R28379_1 (213556_at), score: 0.46 LOC390998similar to hCG1644589 (216589_at), score: -0.46 LOC80054hypothetical LOC80054 (220465_at), score: -0.54 LPAR4lysophosphatidic acid receptor 4 (206960_at), score: 0.45 LPHN1latrophilin 1 (203488_at), score: -0.46 LRRC37A2leucine rich repeat containing 37, member A2 (221740_x_at), score: -0.58 LY6G5Clymphocyte antigen 6 complex, locus G5C (219860_at), score: 1 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: 0.5 MAPK8IP1mitogen-activated protein kinase 8 interacting protein 1 (213013_at), score: -0.46 MARK1MAP/microtubule affinity-regulating kinase 1 (221047_s_at), score: -0.47 MAST4microtubule associated serine/threonine kinase family member 4 (40016_g_at), score: 0.44 MATN3matrilin 3 (206091_at), score: 0.64 MBmyoglobin (204179_at), score: 0.45 MEOX1mesenchyme homeobox 1 (205619_s_at), score: -0.47 MEP1Bmeprin A, beta (207251_at), score: 0.74 MICAL3microtubule associated monoxygenase, calponin and LIM domain containing 3 (212715_s_at), score: 0.52 MPP2membrane protein, palmitoylated 2 (MAGUK p55 subfamily member 2) (213270_at), score: -0.49 MPP3membrane protein, palmitoylated 3 (MAGUK p55 subfamily member 3) (206186_at), score: -0.45 MSR1macrophage scavenger receptor 1 (208423_s_at), score: -0.51 MTMR8myotubularin related protein 8 (220537_at), score: 0.46 MUC5ACmucin 5AC, oligomeric mucus/gel-forming (217182_at), score: -0.55 MUL1mitochondrial E3 ubiquitin ligase 1 (218246_at), score: 0.48 MYL4myosin, light chain 4, alkali; atrial, embryonic (210088_x_at), score: -0.52 NCAM1neural cell adhesion molecule 1 (212843_at), score: -0.47 NCAM2neural cell adhesion molecule 2 (205669_at), score: -0.52 NECAB3N-terminal EF-hand calcium binding protein 3 (210720_s_at), score: -0.62 NOL10nucleolar protein 10 (218591_s_at), score: -0.63 NOVA2neuro-oncological ventral antigen 2 (206477_s_at), score: -0.7 NR1D2nuclear receptor subfamily 1, group D, member 2 (209750_at), score: -0.48 NTRK2neurotrophic tyrosine kinase, receptor, type 2 (207152_at), score: -0.75 NXPH3neurexophilin 3 (221991_at), score: -0.64 OAS32'-5'-oligoadenylate synthetase 3, 100kDa (218400_at), score: 0.47 OR7E156Polfactory receptor, family 7, subfamily E, member 156 pseudogene (222327_x_at), score: -0.47 OR7E47Polfactory receptor, family 7, subfamily E, member 47 pseudogene (216698_x_at), score: -0.53 OVOL2ovo-like 2 (Drosophila) (211778_s_at), score: -0.63 P2RY4pyrimidinergic receptor P2Y, G-protein coupled, 4 (221466_at), score: -0.52 PARVBparvin, beta (37965_at), score: 0.52 PDE1Cphosphodiesterase 1C, calmodulin-dependent 70kDa (207303_at), score: -0.56 PDK3pyruvate dehydrogenase kinase, isozyme 3 (221957_at), score: 0.61 PEG3paternally expressed 3 (209242_at), score: -0.49 PIP5K3phosphatidylinositol-3-phosphate/phosphatidylinositol 5-kinase, type III (213111_at), score: -0.53 PKLRpyruvate kinase, liver and RBC (222078_at), score: -0.45 POU6F1POU class 6 homeobox 1 (205878_at), score: -0.48 PRKCBprotein kinase C, beta (209685_s_at), score: -0.6 PRKCDprotein kinase C, delta (202545_at), score: 0.51 PRR14proline rich 14 (218714_at), score: 0.48 PTP4A3protein tyrosine phosphatase type IVA, member 3 (209695_at), score: -0.46 PTPN6protein tyrosine phosphatase, non-receptor type 6 (206687_s_at), score: -0.57 PTPRRprotein tyrosine phosphatase, receptor type, R (210675_s_at), score: 0.49 PXNpaxillin (211823_s_at), score: 0.56 RAB11FIP3RAB11 family interacting protein 3 (class II) (203933_at), score: 0.52 RAB27BRAB27B, member RAS oncogene family (207018_s_at), score: 0.53 RAG1AP1recombination activating gene 1 activating protein 1 (219125_s_at), score: -0.53 RARRES1retinoic acid receptor responder (tazarotene induced) 1 (221872_at), score: 0.47 RBKSribokinase (219222_at), score: 0.49 RFX3regulatory factor X, 3 (influences HLA class II expression) (207234_at), score: 0.8 RICH2Rho-type GTPase-activating protein RICH2 (215232_at), score: -0.5 RIMS2regulating synaptic membrane exocytosis 2 (215478_at), score: -0.81 RLN1relaxin 1 (211753_s_at), score: -0.49 RNF31ring finger protein 31 (220788_s_at), score: 0.51 RPAINRPA interacting protein (216961_s_at), score: -0.56 RPGRretinitis pigmentosa GTPase regulator (207624_s_at), score: 0.58 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: -0.7 RPS17P5ribosomal protein S17 pseudogene 5 (216348_at), score: -0.58 RRHretinal pigment epithelium-derived rhodopsin homolog (208314_at), score: 0.54 RRP8ribosomal RNA processing 8, methyltransferase, homolog (yeast) (203171_s_at), score: 0.52 S100A14S100 calcium binding protein A14 (218677_at), score: -0.52 SAA4serum amyloid A4, constitutive (207096_at), score: -0.52 SBNO1strawberry notch homolog 1 (Drosophila) (218737_at), score: -0.56 SCAMP4secretory carrier membrane protein 4 (213244_at), score: 0.46 SEC14L5SEC14-like 5 (S. cerevisiae) (210169_at), score: -0.49 SEC61A2Sec61 alpha 2 subunit (S. cerevisiae) (219499_at), score: 0.49 SEPT5septin 5 (209767_s_at), score: -0.63 SERPINA1serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 (211429_s_at), score: -0.68 SETD6SET domain containing 6 (219751_at), score: -0.67 SIRT6sirtuin (silent mating type information regulation 2 homolog) 6 (S. cerevisiae) (219613_s_at), score: 0.51 SLC15A1solute carrier family 15 (oligopeptide transporter), member 1 (211349_at), score: -0.55 SLC16A6solute carrier family 16, member 6 (monocarboxylic acid transporter 7) (207038_at), score: 0.45 SLC19A1solute carrier family 19 (folate transporter), member 1 (211576_s_at), score: -0.46 SLC1A7solute carrier family 1 (glutamate transporter), member 7 (210923_at), score: -0.49 SLC25A16solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 (210686_x_at), score: -0.47 SLC2A8solute carrier family 2 (facilitated glucose transporter), member 8 (218985_at), score: 0.49 SLC37A4solute carrier family 37 (glucose-6-phosphate transporter), member 4 (217289_s_at), score: 0.51 SLC48A1solute carrier family 48 (heme transporter), member 1 (218417_s_at), score: 0.51 SLC4A10solute carrier family 4, sodium bicarbonate transporter, member 10 (206830_at), score: 0.5 SLC5A5solute carrier family 5 (sodium iodide symporter), member 5 (211123_at), score: -0.52 SLC9A3R1solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 1 (201349_at), score: -0.69 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: 0.44 SORL1sortilin-related receptor, L(DLR class) A repeats-containing (203509_at), score: -0.6 SPAG1sperm associated antigen 1 (210117_at), score: 0.46 SPARCL1SPARC-like 1 (hevin) (200795_at), score: -0.53 SPATA1spermatogenesis associated 1 (221057_at), score: -0.55 SPATA6spermatogenesis associated 6 (220298_s_at), score: -0.6 SPG21spastic paraplegia 21 (autosomal recessive, Mast syndrome) (215383_x_at), score: -0.49 SSX3synovial sarcoma, X breakpoint 3 (215881_x_at), score: -0.68 ST7suppression of tumorigenicity 7 (207871_s_at), score: -0.58 SULT1A2sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2 (207122_x_at), score: -0.69 SYN2synapsin II (210247_at), score: -0.61 SYT5synaptotagmin V (206161_s_at), score: -0.75 TAOK2TAO kinase 2 (204986_s_at), score: 0.5 TAS2R14taste receptor, type 2, member 14 (221391_at), score: -0.48 TBX2T-box 2 (40560_at), score: 0.44 TCP10t-complex 10 homolog (mouse) (207503_at), score: -0.51 TGIF2TGFB-induced factor homeobox 2 (216262_s_at), score: -0.65 THNSL1threonine synthase-like 1 (S. cerevisiae) (219800_s_at), score: 0.57 TLE6transducin-like enhancer of split 6 (E(sp1) homolog, Drosophila) (222219_s_at), score: -0.51 TLR3toll-like receptor 3 (206271_at), score: 0.49 TMC5transmembrane channel-like 5 (219580_s_at), score: -0.54 TMEM159transmembrane protein 159 (213272_s_at), score: -0.48 TMEM35transmembrane protein 35 (219685_at), score: 0.6 TMSB4Ythymosin beta 4, Y-linked (206769_at), score: -0.53 TRIM45tripartite motif-containing 45 (219923_at), score: -0.54 TRPM3transient receptor potential cation channel, subfamily M, member 3 (220463_at), score: 0.55 TULP2tubby like protein 2 (206733_at), score: -0.49 TYRO3TYRO3 protein tyrosine kinase (211432_s_at), score: 0.46 UBTD1ubiquitin domain containing 1 (219172_at), score: 0.59 VAMP8vesicle-associated membrane protein 8 (endobrevin) (202546_at), score: -0.68 VCPIP1valosin containing protein (p97)/p47 complex interacting protein 1 (219810_at), score: -0.69 VEGFBvascular endothelial growth factor B (203683_s_at), score: 0.51 VPS37Bvacuolar protein sorting 37 homolog B (S. cerevisiae) (221704_s_at), score: 0.5 WDR4WD repeat domain 4 (221632_s_at), score: -0.49 WFDC2WAP four-disulfide core domain 2 (203892_at), score: -0.59 WNT2Bwingless-type MMTV integration site family, member 2B (206459_s_at), score: -0.63 YPEL1yippee-like 1 (Drosophila) (206063_x_at), score: -0.46 ZBTB7Azinc finger and BTB domain containing 7A (219186_at), score: -0.5 ZDHHC14zinc finger, DHHC-type containing 14 (219247_s_at), score: -0.68 ZFYVE26zinc finger, FYVE domain containing 26 (37943_at), score: -0.46 ZHX3zinc fingers and homeoboxes 3 (212545_s_at), score: 0.44 ZNF124zinc finger protein 124 (206928_at), score: -0.54 ZNF254zinc finger protein 254 (206862_at), score: 0.45 ZNF281zinc finger protein 281 (218401_s_at), score: -0.46 ZNF323zinc finger protein 323 (222016_s_at), score: 0.49 ZNF362zinc finger protein 362 (214915_at), score: -0.61 ZNF518Azinc finger protein 518A (204291_at), score: 0.57 ZNF614zinc finger protein 614 (220721_at), score: -0.46 ZNF667zinc finger protein 667 (207120_at), score: -0.52 ZNF814zinc finger protein 814 (60794_f_at), score: 0.59
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485791.cel | 8 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486171.cel | 27 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486071.cel | 22 | 6 | Cycle | hgu133a2 | none | Cycle 1 |