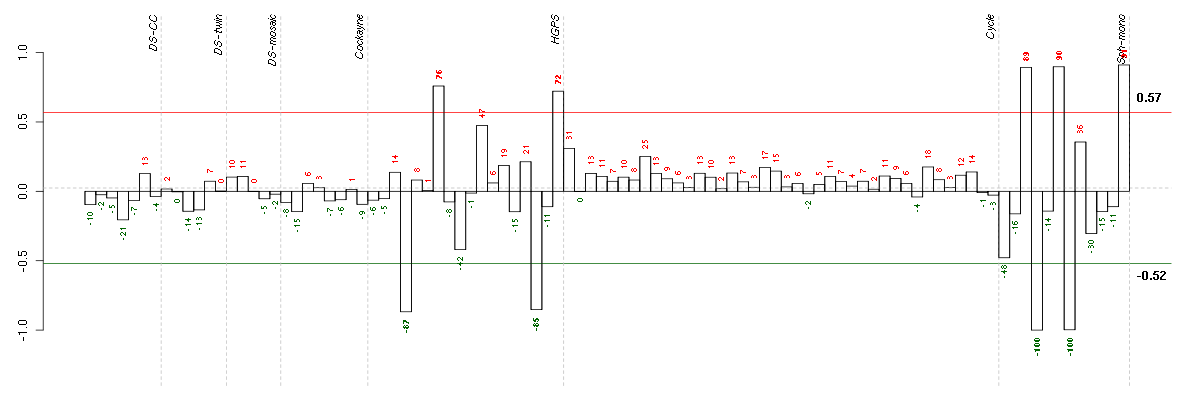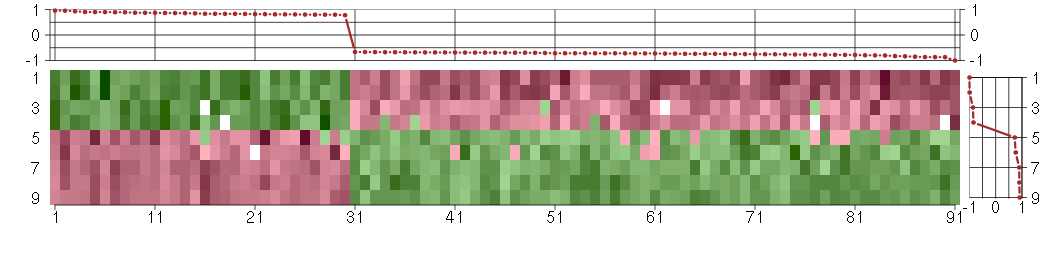

Under-expression is coded with green,
over-expression with red color.

immune system process
Any process involved in the development or functioning of the immune system, an organismal system for calibrated responses to potential internal or invasive threats.
signal transduction
The cascade of processes by which a signal interacts with a receptor, causing a change in the level or activity of a second messenger or other downstream target, and ultimately effecting a change in the functioning of the cell.
cell communication
Any process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
cell surface receptor linked signal transduction
Any series of molecular signals initiated by the binding of an extracellular ligand to a receptor on the surface of the target cell.
enzyme linked receptor protein signaling pathway
Any series of molecular signals initiated by the binding of an extracellular ligand to a receptor on the surface of the target cell, where the receptor possesses catalytic activity or is closely associated with an enzyme such as a protein kinase.
transmembrane receptor protein tyrosine kinase signaling pathway
The series of molecular signals generated as a consequence of a transmembrane receptor tyrosine kinase binding to its physiological ligand.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
regulation of signal transduction
Any process that modulates the frequency, rate or extent of signal transduction.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of cell communication
Any process that modulates the frequency, rate or extent of cell communication. Cell communication is the process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
antigen processing and presentation
The process by which an antigen-presenting cell expresses antigen (peptide or lipid) on its cell surface in association with an MHC protein complex.
antigen processing and presentation of peptide antigen
The process by which an antigen-presenting cell expresses peptide antigen in association with an MHC protein complex on its cell surface, including proteolysis and transport steps for the peptide antigen both prior to and following assembly with the MHC protein complex. The peptide antigen is typically, but not always, processed from an endogenous or exogenous protein.
nerve growth factor receptor signaling pathway
The series of molecular signals generated as a consequence of the nerve growth factor receptor binding to one of its physiological ligands.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of nerve growth factor receptor signaling pathway
Any process that modulates the frequency, rate or extent of the activity of the nerve growth factor receptor signaling pathway.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
all
This term is the most general term possible
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
signal transduction
The cascade of processes by which a signal interacts with a receptor, causing a change in the level or activity of a second messenger or other downstream target, and ultimately effecting a change in the functioning of the cell.
regulation of cell communication
Any process that modulates the frequency, rate or extent of cell communication. Cell communication is the process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
regulation of signal transduction
Any process that modulates the frequency, rate or extent of signal transduction.
regulation of nerve growth factor receptor signaling pathway
Any process that modulates the frequency, rate or extent of the activity of the nerve growth factor receptor signaling pathway.


molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
signal transducer activity
Mediates the transfer of a signal from the outside to the inside of a cell by means other than the introduction of the signal molecule itself into the cell.
receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity.
transmembrane receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity, and spanning to the membrane of either the cell or an organelle.
MHC class I receptor activity
Combining with an MHC class I protein complex to initiate a change in cellular activity. Class I here refers to classical class I molecules.
molecular transducer activity
The molecular function that accepts an input of one form and creates an output of a different form.
all
This term is the most general term possible
ABHD2abhydrolase domain containing 2 (221815_at), score: -0.78 ADIPOR1adiponectin receptor 1 (217748_at), score: -0.85 AGTangiotensinogen (serpin peptidase inhibitor, clade A, member 8) (202834_at), score: -0.72 ALDH1A1aldehyde dehydrogenase 1 family, member A1 (212224_at), score: 0.89 ALDOAP2aldolase A, fructose-bisphosphate pseudogene 2 (211617_at), score: -0.67 ANXA3annexin A3 (209369_at), score: 0.82 APPamyloid beta (A4) precursor protein (200602_at), score: -0.67 ARHGAP29Rho GTPase activating protein 29 (203910_at), score: 0.87 ARL4CADP-ribosylation factor-like 4C (202207_at), score: -0.72 BMP7bone morphogenetic protein 7 (209590_at), score: -0.68 C17orf86chromosome 17 open reading frame 86 (221621_at), score: -0.72 C20orf12chromosome 20 open reading frame 12 (219951_s_at), score: -0.74 C6orf162chromosome 6 open reading frame 162 (213314_at), score: -0.69 CDC14BCDC14 cell division cycle 14 homolog B (S. cerevisiae) (208022_s_at), score: -0.75 CDKN1Ccyclin-dependent kinase inhibitor 1C (p57, Kip2) (213183_s_at), score: -0.72 CELSR3cadherin, EGF LAG seven-pass G-type receptor 3 (flamingo homolog, Drosophila) (40020_at), score: -0.72 CFHcomplement factor H (213800_at), score: 0.8 CHRDL1chordin-like 1 (209763_at), score: 0.95 CUGBP2CUG triplet repeat, RNA binding protein 2 (202158_s_at), score: 0.87 CYP2B6cytochrome P450, family 2, subfamily B, polypeptide 6 (217133_x_at), score: -0.78 DKFZP586I1420hypothetical protein DKFZp586I1420 (213546_at), score: -0.72 DSTYKdual serine/threonine and tyrosine protein kinase (214663_at), score: -0.73 DUSP3dual specificity phosphatase 3 (201536_at), score: -0.69 EN1engrailed homeobox 1 (220559_at), score: -0.75 FAM134Cfamily with sequence similarity 134, member C (212697_at), score: -0.75 FBXO41F-box protein 41 (44040_at), score: -0.78 FCARFc fragment of IgA, receptor for (211305_x_at), score: -0.77 FUCA1fucosidase, alpha-L- 1, tissue (202838_at), score: -0.69 FUT8fucosyltransferase 8 (alpha (1,6) fucosyltransferase) (203988_s_at), score: 0.81 FZD5frizzled homolog 5 (Drosophila) (221245_s_at), score: -0.75 GJA1gap junction protein, alpha 1, 43kDa (201667_at), score: 0.9 GPR126G protein-coupled receptor 126 (213094_at), score: 0.9 GREM1gremlin 1, cysteine knot superfamily, homolog (Xenopus laevis) (218469_at), score: 0.83 HLA-Amajor histocompatibility complex, class I, A (213932_x_at), score: -0.69 HLA-Bmajor histocompatibility complex, class I, B (211911_x_at), score: -0.79 HLA-Cmajor histocompatibility complex, class I, C (211799_x_at), score: -0.69 HPS4Hermansky-Pudlak syndrome 4 (54037_at), score: -0.7 IFI30interferon, gamma-inducible protein 30 (201422_at), score: -0.74 IGSF3immunoglobulin superfamily, member 3 (202421_at), score: -0.72 JAM3junctional adhesion molecule 3 (212813_at), score: 0.93 JARID2jumonji, AT rich interactive domain 2 (203297_s_at), score: -0.72 KCNAB1potassium voltage-gated channel, shaker-related subfamily, beta member 1 (210078_s_at), score: -0.76 KCNJ14potassium inwardly-rectifying channel, subfamily J, member 14 (220776_at), score: -0.75 LHFPlipoma HMGIC fusion partner (218656_s_at), score: 0.81 LOC729034similar to puromycin sensitive aminopeptidase (214107_x_at), score: -0.68 LOC91316glucuronidase, beta/ immunoglobulin lambda-like polypeptide 1 pseudogene (215816_at), score: -0.7 LTBP2latent transforming growth factor beta binding protein 2 (204682_at), score: 0.81 MAFGv-maf musculoaponeurotic fibrosarcoma oncogene homolog G (avian) (204970_s_at), score: -0.82 MAP2K7mitogen-activated protein kinase kinase 7 (216206_x_at), score: -0.72 MAPRE3microtubule-associated protein, RP/EB family, member 3 (214270_s_at), score: -0.67 MPDZmultiple PDZ domain protein (213306_at), score: 0.85 MTUS1mitochondrial tumor suppressor 1 (212096_s_at), score: -0.8 MYH10myosin, heavy chain 10, non-muscle (212372_at), score: 0.96 NACA2nascent polypeptide-associated complex alpha subunit 2 (222224_at), score: -0.74 NRN1neuritin 1 (218625_at), score: -0.67 PCGF2polycomb group ring finger 2 (203793_x_at), score: -0.71 PFTK1PFTAIRE protein kinase 1 (204604_at), score: 0.86 PIGNphosphatidylinositol glycan anchor biosynthesis, class N (219048_at), score: 0.83 PMS2CLPMS2 C-terminal like pseudogene (221206_at), score: -0.77 PPARGC1Aperoxisome proliferator-activated receptor gamma, coactivator 1 alpha (219195_at), score: -0.87 PSMD5proteasome (prosome, macropain) 26S subunit, non-ATPase, 5 (203447_at), score: 0.83 PVT1Pvt1 oncogene (non-protein coding) (216249_at), score: -0.77 PYGLphosphorylase, glycogen, liver (202990_at), score: 0.87 RHBDL2rhomboid, veinlet-like 2 (Drosophila) (219489_s_at), score: 0.86 ROBO1roundabout, axon guidance receptor, homolog 1 (Drosophila) (213194_at), score: 0.83 SAPS2SAPS domain family, member 2 (202792_s_at), score: -0.67 SETD1ASET domain containing 1A (213202_at), score: -0.87 SIM2single-minded homolog 2 (Drosophila) (206558_at), score: -1 SLC16A6solute carrier family 16, member 6 (monocarboxylic acid transporter 7) (207038_at), score: -0.68 SLC22A17solute carrier family 22, member 17 (218675_at), score: 0.8 SLC23A2solute carrier family 23 (nucleobase transporters), member 2 (209236_at), score: -0.74 SLC24A1solute carrier family 24 (sodium/potassium/calcium exchanger), member 1 (206081_at), score: 0.8 SPATA1spermatogenesis associated 1 (221057_at), score: -0.74 STX3syntaxin 3 (209238_at), score: -0.7 SULF1sulfatase 1 (212353_at), score: 0.83 THBS2thrombospondin 2 (203083_at), score: 0.86 TMEM80transmembrane protein 80 (221951_at), score: -0.72 TPBGtrophoblast glycoprotein (203476_at), score: 0.82 TSPYL5TSPY-like 5 (213122_at), score: 0.89 TUBB6tubulin, beta 6 (209191_at), score: 0.78 ULK1unc-51-like kinase 1 (C. elegans) (209333_at), score: -0.77 UPP1uridine phosphorylase 1 (203234_at), score: -0.68 USF2upstream transcription factor 2, c-fos interacting (214879_x_at), score: -0.8 VAMP2vesicle-associated membrane protein 2 (synaptobrevin 2) (201557_at), score: -0.86 ZDHHC7zinc finger, DHHC-type containing 7 (218606_at), score: -0.69 ZNF365zinc finger protein 365 (206448_at), score: 0.8 ZNF440zinc finger protein 440 (215892_at), score: -0.73 ZNF639zinc finger protein 639 (218413_s_at), score: -0.84 ZNF749zinc finger protein 749 (215289_at), score: -0.72 ZNF816Azinc finger protein 816A (217541_x_at), score: -0.69
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-4219-raw-cel-1311956275.cel | 8 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956398.cel | 12 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-3860-raw-cel-1561690231.cel | 4 | 5 | HGPS | hgu133a | HGPS | AG10750 |
| E-GEOD-3860-raw-cel-1561690432.cel | 16 | 5 | HGPS | hgu133a | HGPS | AG10750 |
| E-GEOD-3860-raw-cel-1561690480.cel | 18 | 5 | HGPS | hgu133a | HGPS | AG11498 |
| E-GEOD-3860-raw-cel-1561690272.cel | 7 | 5 | HGPS | hgu133a | HGPS | AG11498 |
| E-GEOD-4219-raw-cel-1311956178.cel | 6 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956358.cel | 10 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956824.cel | 24 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |