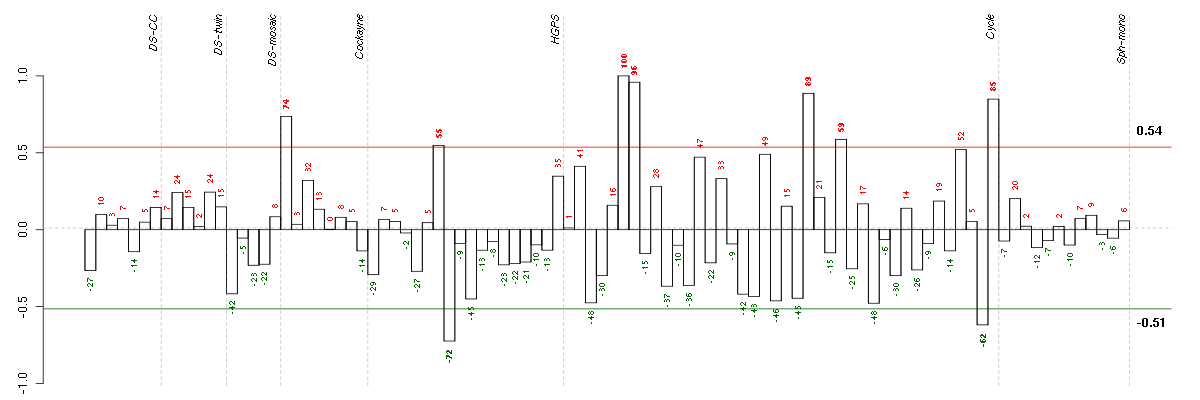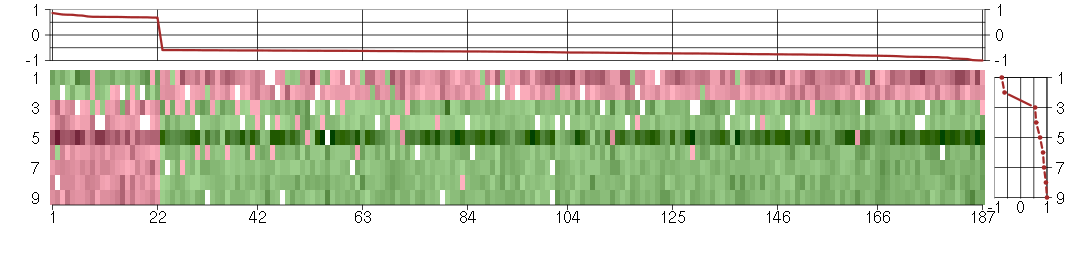

Under-expression is coded with green,
over-expression with red color.



ABCD1ATP-binding cassette, sub-family D (ALD), member 1 (205142_x_at), score: -0.69 ADCK2aarF domain containing kinase 2 (221893_s_at), score: -0.63 ADCY3adenylate cyclase 3 (209321_s_at), score: -0.82 AGPAT11-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) (215535_s_at), score: -0.73 AGRNagrin (212285_s_at), score: -0.62 ANAPC2anaphase promoting complex subunit 2 (218555_at), score: -0.61 ARFGAP1ADP-ribosylation factor GTPase activating protein 1 (217888_s_at), score: -0.66 ARSAarylsulfatase A (204443_at), score: -0.72 ATN1atrophin 1 (40489_at), score: -0.85 B3GAT3beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) (203452_at), score: -0.69 BAP1BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) (201419_at), score: -0.65 BAT2HLA-B associated transcript 2 (212081_x_at), score: -0.65 BAT2LHLA-B associated transcript 2-like (212068_s_at), score: -0.64 BIN1bridging integrator 1 (210201_x_at), score: -0.94 BRCA2breast cancer 2, early onset (208368_s_at), score: 0.68 BRD9bromodomain containing 9 (220155_s_at), score: -0.72 BTBD2BTB (POZ) domain containing 2 (207722_s_at), score: -0.78 C14orf106chromosome 14 open reading frame 106 (206500_s_at), score: 0.79 C16orf57chromosome 16 open reading frame 57 (218060_s_at), score: -0.69 C16orf58chromosome 16 open reading frame 58 (217891_at), score: -0.59 C19orf22chromosome 19 open reading frame 22 (221764_at), score: -0.66 C20orf20chromosome 20 open reading frame 20 (218586_at), score: -0.62 C22orf29chromosome 22 open reading frame 29 (219560_at), score: -0.64 CABIN1calcineurin binding protein 1 (37652_at), score: -0.64 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (217849_s_at), score: -0.85 CDC42EP1CDC42 effector protein (Rho GTPase binding) 1 (204693_at), score: -0.72 CDR2Lcerebellar degeneration-related protein 2-like (213230_at), score: -0.86 CENPC1centromere protein C 1 (204739_at), score: 0.71 CENPQcentromere protein Q (219294_at), score: 0.71 CEP192centrosomal protein 192kDa (218827_s_at), score: 0.79 CEP57centrosomal protein 57kDa (203491_s_at), score: 0.71 CHPFchondroitin polymerizing factor (202175_at), score: -0.6 CHST2carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2 (203921_at), score: -0.63 CHST3carbohydrate (chondroitin 6) sulfotransferase 3 (209834_at), score: -0.7 CICcapicua homolog (Drosophila) (212784_at), score: -0.75 CIZ1CDKN1A interacting zinc finger protein 1 (205516_x_at), score: -0.75 CLIP2CAP-GLY domain containing linker protein 2 (211031_s_at), score: -0.75 CLIP3CAP-GLY domain containing linker protein 3 (212358_at), score: -0.61 CNN2calponin 2 (201605_x_at), score: -0.8 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: -0.61 CORO2Bcoronin, actin binding protein, 2B (209789_at), score: -0.7 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: -0.77 DEXIdexamethasone-induced transcript (203733_at), score: -0.64 DMPKdystrophia myotonica-protein kinase (37996_s_at), score: -0.62 DMWDdystrophia myotonica, WD repeat containing (33768_at), score: -0.8 DOK4docking protein 4 (209691_s_at), score: -0.76 DSC3desmocollin 3 (206032_at), score: -0.76 FAM134Cfamily with sequence similarity 134, member C (212697_at), score: -0.71 FASNfatty acid synthase (212218_s_at), score: -0.83 FBXL11F-box and leucine-rich repeat protein 11 (208989_s_at), score: -0.65 FBXO17F-box protein 17 (220233_at), score: -0.84 FCF1FCF1 small subunit (SSU) processome component homolog (S. cerevisiae) (219927_at), score: 0.71 FHOD1formin homology 2 domain containing 1 (218530_at), score: -0.61 FKSG2apoptosis inhibitor (208588_at), score: 0.71 FLJ12529pre-mRNA cleavage factor I, 59 kDa subunit (217866_at), score: -0.74 FOXK2forkhead box K2 (203064_s_at), score: -1 FURINfurin (paired basic amino acid cleaving enzyme) (201945_at), score: -0.7 GFPT2glutamine-fructose-6-phosphate transaminase 2 (205100_at), score: -0.6 GOLGA3golgi autoantigen, golgin subfamily a, 3 (202106_at), score: -0.72 GOLPH3golgi phosphoprotein 3 (coat-protein) (217803_at), score: -0.62 GP1BBglycoprotein Ib (platelet), beta polypeptide (206655_s_at), score: -0.72 GRINAglutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) (212090_at), score: -0.78 GTPBP1GTP binding protein 1 (219357_at), score: -0.62 HDAC5histone deacetylase 5 (202455_at), score: -0.6 HECTD3HECT domain containing 3 (218632_at), score: -0.69 HSF1heat shock transcription factor 1 (202344_at), score: -0.64 HSPA12Aheat shock 70kDa protein 12A (214434_at), score: -0.69 IDUAiduronidase, alpha-L- (205059_s_at), score: -0.82 JARID1Cjumonji, AT rich interactive domain 1C (202383_at), score: -0.62 KCTD13potassium channel tetramerisation domain containing 13 (45653_at), score: -0.87 KIAA0317KIAA0317 (202128_at), score: -0.59 KIAA1009KIAA1009 (206005_s_at), score: 0.8 KIAA1305KIAA1305 (220911_s_at), score: -0.66 KLHL26kelch-like 26 (Drosophila) (219354_at), score: -0.59 KPNA6karyopherin alpha 6 (importin alpha 7) (212101_at), score: -0.72 LEPREL2leprecan-like 2 (204854_at), score: -0.62 LOC339047hypothetical protein LOC339047 (221501_x_at), score: -0.68 LPPR2lipid phosphate phosphatase-related protein type 2 (218509_at), score: -0.64 LRFN4leucine rich repeat and fibronectin type III domain containing 4 (219491_at), score: -0.59 LRIG1leucine-rich repeats and immunoglobulin-like domains 1 (211596_s_at), score: -0.64 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: -0.93 MAP2K3mitogen-activated protein kinase kinase 3 (207667_s_at), score: -0.7 MAP3K11mitogen-activated protein kinase kinase kinase 11 (203652_at), score: -0.69 MAP3K5mitogen-activated protein kinase kinase kinase 5 (203837_at), score: 0.7 MAP4microtubule-associated protein 4 (200836_s_at), score: -0.64 MAP7D1MAP7 domain containing 1 (217943_s_at), score: -0.81 MAST2microtubule associated serine/threonine kinase 2 (211593_s_at), score: -0.62 MAST4microtubule associated serine/threonine kinase family member 4 (40016_g_at), score: -0.66 MC4Rmelanocortin 4 receptor (221467_at), score: 0.7 MED15mediator complex subunit 15 (222175_s_at), score: -0.6 MGRN1mahogunin, ring finger 1 (212576_at), score: -0.61 MICAL3microtubule associated monoxygenase, calponin and LIM domain containing 3 (212715_s_at), score: -0.63 MINK1misshapen-like kinase 1 (zebrafish) (214246_x_at), score: -0.68 MLL4myeloid/lymphoid or mixed-lineage leukemia 4 (203419_at), score: -0.72 MUL1mitochondrial E3 ubiquitin ligase 1 (218246_at), score: -0.63 MYO9Bmyosin IXB (217297_s_at), score: -0.65 MYST3MYST histone acetyltransferase (monocytic leukemia) 3 (202423_at), score: -0.62 NCDNneurochondrin (209556_at), score: -0.79 NCKAP1NCK-associated protein 1 (207738_s_at), score: 0.7 NCOR2nuclear receptor co-repressor 2 (207760_s_at), score: -0.69 NINJ1ninjurin 1 (203045_at), score: -0.62 NLGN1neuroligin 1 (205893_at), score: 0.76 NOL9nucleolar protein 9 (218754_at), score: -0.63 NPIPnuclear pore complex interacting protein (204538_x_at), score: -0.63 NUDT11nudix (nucleoside diphosphate linked moiety X)-type motif 11 (219855_at), score: -0.62 OS9osteosarcoma amplified 9, endoplasmic reticulum associated protein (200714_x_at), score: -0.6 PARVBparvin, beta (37965_at), score: -0.74 PCDHG@protocadherin gamma cluster (215836_s_at), score: -0.77 PCDHGA1protocadherin gamma subfamily A, 1 (209079_x_at), score: -0.94 PCGF2polycomb group ring finger 2 (203793_x_at), score: -0.73 PDE4Aphosphodiesterase 4A, cAMP-specific (phosphodiesterase E2 dunce homolog, Drosophila) (204735_at), score: -0.61 PDLIM4PDZ and LIM domain 4 (211564_s_at), score: -0.71 PEX26peroxisomal biogenesis factor 26 (219180_s_at), score: -0.6 PHC1polyhomeotic homolog 1 (Drosophila) (218338_at), score: -0.66 PHF20PHD finger protein 20 (209422_at), score: 0.69 PIP5K1Cphosphatidylinositol-4-phosphate 5-kinase, type I, gamma (212518_at), score: -0.99 PKD1polycystic kidney disease 1 (autosomal dominant) (202328_s_at), score: -0.65 PKN1protein kinase N1 (202161_at), score: -0.76 PLAGL2pleiomorphic adenoma gene-like 2 (202925_s_at), score: -0.74 PLCXD1phosphatidylinositol-specific phospholipase C, X domain containing 1 (218951_s_at), score: -0.6 PNPLA6patatin-like phospholipase domain containing 6 (203718_at), score: -0.73 POLR2Apolymerase (RNA) II (DNA directed) polypeptide A, 220kDa (202725_at), score: -0.75 POLRMTpolymerase (RNA) mitochondrial (DNA directed) (203782_s_at), score: -0.59 POM121POM121 membrane glycoprotein (rat) (212178_s_at), score: -0.78 POM121CPOM121 membrane glycoprotein C (213360_s_at), score: -0.63 POMZP3POM (POM121 homolog, rat) and ZP3 fusion (204148_s_at), score: -0.76 PPM1Fprotein phosphatase 1F (PP2C domain containing) (203063_at), score: -0.64 PRKCDprotein kinase C, delta (202545_at), score: -0.61 PRR5proline rich 5 (renal) (47069_at), score: -0.62 PTK7PTK7 protein tyrosine kinase 7 (207011_s_at), score: -0.67 PTPN9protein tyrosine phosphatase, non-receptor type 9 (202958_at), score: -0.72 PTPROprotein tyrosine phosphatase, receptor type, O (211600_at), score: -0.71 RAB22ARAB22A, member RAS oncogene family (218360_at), score: -0.61 RAD17RAD17 homolog (S. pombe) (207405_s_at), score: 0.68 RAD54L2RAD54-like 2 (S. cerevisiae) (213205_s_at), score: -0.82 RBM15BRNA binding motif protein 15B (202689_at), score: -0.79 REREarginine-glutamic acid dipeptide (RE) repeats (200940_s_at), score: -0.74 RGS2regulator of G-protein signaling 2, 24kDa (202388_at), score: -0.66 RNF187ring finger protein 187 (212155_at), score: -0.73 RNF24ring finger protein 24 (204669_s_at), score: -0.61 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: 0.86 RPRD2regulation of nuclear pre-mRNA domain containing 2 (212553_at), score: -0.65 RPS17P5ribosomal protein S17 pseudogene 5 (216348_at), score: 0.73 SCAMP1secretory carrier membrane protein 1 (206667_s_at), score: 0.77 SCAMP4secretory carrier membrane protein 4 (213244_at), score: -0.93 SDC3syndecan 3 (202898_at), score: -0.62 SENP5SUMO1/sentrin specific peptidase 5 (213184_at), score: -0.72 SF1splicing factor 1 (208313_s_at), score: -0.75 SHBSrc homology 2 domain containing adaptor protein B (204657_s_at), score: -0.64 SIN3BSIN3 homolog B, transcription regulator (yeast) (209352_s_at), score: -0.65 SIRT6sirtuin (silent mating type information regulation 2 homolog) 6 (S. cerevisiae) (219613_s_at), score: -0.7 SLC25A22solute carrier family 25 (mitochondrial carrier: glutamate), member 22 (218725_at), score: -0.61 SLC31A2solute carrier family 31 (copper transporters), member 2 (204204_at), score: 0.83 SLC6A8solute carrier family 6 (neurotransmitter transporter, creatine), member 8 (202219_at), score: -0.69 SLC9A1solute carrier family 9 (sodium/hydrogen exchanger), member 1 (209453_at), score: -0.66 SMARCD1SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 (209518_at), score: -0.61 SMG7Smg-7 homolog, nonsense mediated mRNA decay factor (C. elegans) (201794_s_at), score: -0.81 SMOXspermine oxidase (210357_s_at), score: -0.61 SMYD5SMYD family member 5 (209516_at), score: -0.73 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: -0.81 SORBS3sorbin and SH3 domain containing 3 (209253_at), score: -0.6 SPRY4sprouty homolog 4 (Drosophila) (221489_s_at), score: -0.7 SSBP3single stranded DNA binding protein 3 (217991_x_at), score: -0.89 STRN4striatin, calmodulin binding protein 4 (217903_at), score: -0.86 SULT1B1sulfotransferase family, cytosolic, 1B, member 1 (207601_at), score: 0.69 SUPT6Hsuppressor of Ty 6 homolog (S. cerevisiae) (208831_x_at), score: -0.76 TAPBPTAP binding protein (tapasin) (208829_at), score: -0.86 TBC1D10BTBC1 domain family, member 10B (220947_s_at), score: -0.67 TERF2telomeric repeat binding factor 2 (203611_at), score: -0.67 TLE3transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) (206472_s_at), score: -0.62 TMEM149transmembrane protein 149 (219690_at), score: 0.71 TOP3Atopoisomerase (DNA) III alpha (214300_s_at), score: 0.7 TRIM21tripartite motif-containing 21 (204804_at), score: -0.59 TSC2tuberous sclerosis 2 (215735_s_at), score: -0.64 TSKUtsukushin (218245_at), score: -0.98 TSSC4tumor suppressing subtransferable candidate 4 (218612_s_at), score: -0.63 TXLNAtaxilin alpha (212300_at), score: -0.77 UBE3Aubiquitin protein ligase E3A (213291_s_at), score: -0.59 UBTD1ubiquitin domain containing 1 (219172_at), score: -0.76 VAC14Vac14 homolog (S. cerevisiae) (218169_at), score: -0.65 VEGFBvascular endothelial growth factor B (203683_s_at), score: -0.77 VPS37Bvacuolar protein sorting 37 homolog B (S. cerevisiae) (221704_s_at), score: -0.89 WDR42AWD repeat domain 42A (202249_s_at), score: -0.81 WDR6WD repeat domain 6 (217734_s_at), score: -0.72 XAB2XPA binding protein 2 (218110_at), score: -0.87 ZNF282zinc finger protein 282 (212892_at), score: -0.67 ZNF580zinc finger protein 580 (220748_s_at), score: -0.76
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-3860-raw-cel-1561690304.cel | 8 | 5 | HGPS | hgu133a | none | GMO8398C |
| E-TABM-263-raw-cel-1515486411.cel | 39 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690272.cel | 7 | 5 | HGPS | hgu133a | HGPS | AG11498 |
| E-TABM-263-raw-cel-1515486151.cel | 26 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3407-raw-cel-1437949557.cel | 1 | 4 | Cockayne | hgu133a | CS | eGFP |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |