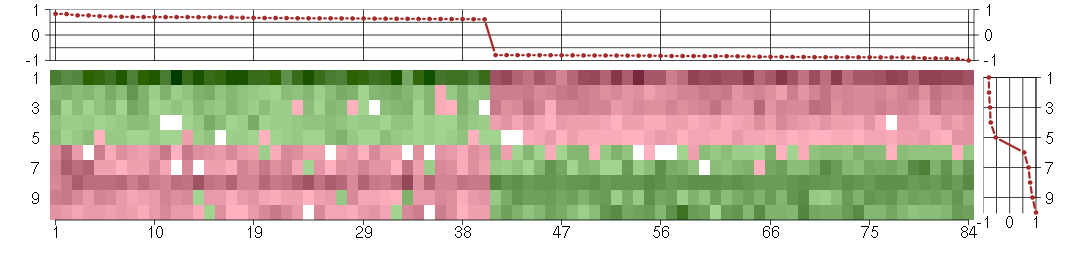

Under-expression is coded with green,
over-expression with red color.


intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
cytoplasm
All of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
secretory granule
A small subcellular vesicle, surrounded by a membrane, that is formed from the Golgi apparatus and contains a highly concentrated protein destined for secretion. Secretory granules move towards the periphery of the cell, their membranes fuse with the cell membrane, and their protein load is exteriorized. Processing of the contained protein may take place in secretory granules.
cytoplasmic membrane-bounded vesicle
A membrane-bounded vesicle found in the cytoplasm of the cell.
cytoplasmic vesicle
A vesicle formed of membrane or protein, found in the cytoplasm of a cell.
vesicle
Any small, fluid-filled, spherical organelle enclosed by membrane or protein.
membrane-bounded vesicle
Any small, fluid-filled, spherical organelle enclosed by a lipid bilayer.
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
cytoplasmic vesicle
A vesicle formed of membrane or protein, found in the cytoplasm of a cell.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
cytoplasmic membrane-bounded vesicle
A membrane-bounded vesicle found in the cytoplasm of the cell.
cytoplasmic membrane-bounded vesicle
A membrane-bounded vesicle found in the cytoplasm of the cell.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
cytoplasmic vesicle
A vesicle formed of membrane or protein, found in the cytoplasm of a cell.

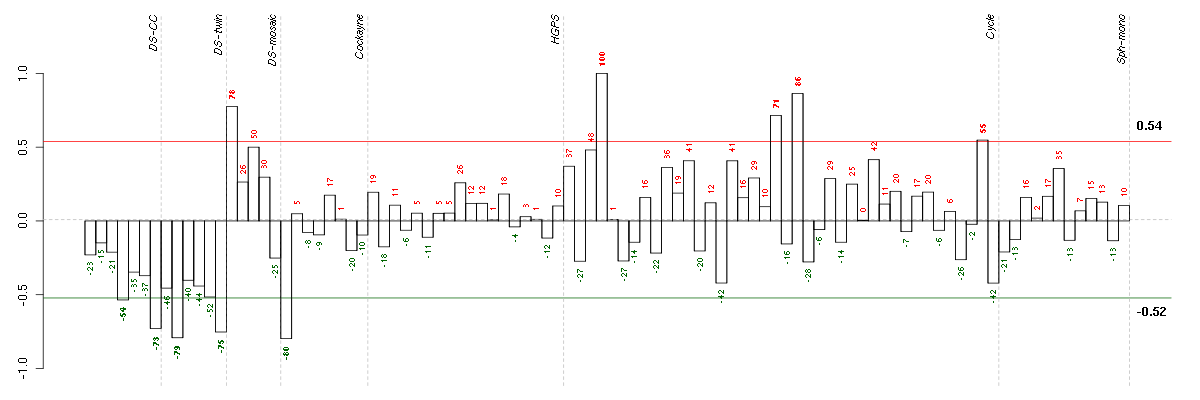
ABLIM3actin binding LIM protein family, member 3 (205730_s_at), score: 0.65 ACRV1acrosomal vesicle protein 1 (207969_x_at), score: -0.8 AIPL1aryl hydrocarbon receptor interacting protein-like 1 (219977_at), score: -0.86 ARSAarylsulfatase A (204443_at), score: 0.69 BAT2LHLA-B associated transcript 2-like (212068_s_at), score: 0.67 BCANbrevican (91920_at), score: -0.79 BCL7AB-cell CLL/lymphoma 7A (203795_s_at), score: 0.77 BTNL3butyrophilin-like 3 (217207_s_at), score: -0.87 C1orf61chromosome 1 open reading frame 61 (205103_at), score: -0.86 C1orf69chromosome 1 open reading frame 69 (215490_at), score: -0.85 C8orf17chromosome 8 open reading frame 17 (208266_at), score: -0.92 C8orf60chromosome 8 open reading frame 60 (220712_at), score: -0.84 C9orf167chromosome 9 open reading frame 167 (219620_x_at), score: 0.73 CALB1calbindin 1, 28kDa (205626_s_at), score: -0.81 CAMK1calcium/calmodulin-dependent protein kinase I (204392_at), score: 0.66 CD79BCD79b molecule, immunoglobulin-associated beta (205297_s_at), score: -0.79 CHST4carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 4 (220446_s_at), score: -0.92 CLDN1claudin 1 (218182_s_at), score: 0.67 COL11A2collagen, type XI, alpha 2 (216993_s_at), score: -0.94 DEAF1deformed epidermal autoregulatory factor 1 (Drosophila) (209407_s_at), score: 0.64 DNAJB5DnaJ (Hsp40) homolog, subfamily B, member 5 (212817_at), score: 0.66 DOK4docking protein 4 (209691_s_at), score: 0.63 DSPPdentin sialophosphoprotein (221681_s_at), score: -0.87 EDARectodysplasin A receptor (220048_at), score: -0.89 FGL1fibrinogen-like 1 (205305_at), score: -0.81 FHOD1formin homology 2 domain containing 1 (218530_at), score: 0.74 GABRR1gamma-aminobutyric acid (GABA) receptor, rho 1 (206525_at), score: -0.83 GNG4guanine nucleotide binding protein (G protein), gamma 4 (205184_at), score: -0.81 GP1BAglycoprotein Ib (platelet), alpha polypeptide (207389_at), score: -0.81 GPR32G protein-coupled receptor 32 (221469_at), score: -0.83 GPX3glutathione peroxidase 3 (plasma) (214091_s_at), score: 0.64 GTPBP1GTP binding protein 1 (219357_at), score: 0.65 HAB1B1 for mucin (215778_x_at), score: -0.83 HAMPhepcidin antimicrobial peptide (220491_at), score: -0.83 HHEXhematopoietically expressed homeobox (215933_s_at), score: 0.63 HLA-Emajor histocompatibility complex, class I, E (200904_at), score: 0.71 HSF1heat shock transcription factor 1 (202344_at), score: 0.71 HTR1E5-hydroxytryptamine (serotonin) receptor 1E (207404_s_at), score: -0.87 IDUAiduronidase, alpha-L- (205059_s_at), score: 0.83 ITGALintegrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide) (213475_s_at), score: -0.82 KLHL26kelch-like 26 (Drosophila) (219354_at), score: 0.65 LEPREL2leprecan-like 2 (204854_at), score: 0.65 LRP5low density lipoprotein receptor-related protein 5 (209468_at), score: 0.65 MADCAM1mucosal vascular addressin cell adhesion molecule 1 (208037_s_at), score: -0.8 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: 0.7 MAP3K6mitogen-activated protein kinase kinase kinase 6 (219278_at), score: 0.63 MEOX1mesenchyme homeobox 1 (205619_s_at), score: -0.85 NACAP1nascent-polypeptide-associated complex alpha polypeptide pseudogene 1 (211445_x_at), score: -0.79 NFKB1nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 (209239_at), score: 0.7 NINJ1ninjurin 1 (203045_at), score: 0.64 OPRL1opiate receptor-like 1 (206564_at), score: -0.82 OR7E24olfactory receptor, family 7, subfamily E, member 24 (215463_at), score: -0.8 OR7E47Polfactory receptor, family 7, subfamily E, member 47 pseudogene (216698_x_at), score: -0.79 PECAM1platelet/endothelial cell adhesion molecule (208982_at), score: -1 PGCprogastricsin (pepsinogen C) (205261_at), score: -0.79 PHF7PHD finger protein 7 (215622_x_at), score: -0.87 PIP5K1Cphosphatidylinositol-4-phosphate 5-kinase, type I, gamma (212518_at), score: 0.7 PKN1protein kinase N1 (202161_at), score: 0.63 PKP2plakophilin 2 (207717_s_at), score: 0.7 PRKCDprotein kinase C, delta (202545_at), score: 0.82 PTGDSprostaglandin D2 synthase 21kDa (brain) (212187_x_at), score: 0.71 PTPN6protein tyrosine phosphatase, non-receptor type 6 (206687_s_at), score: -0.86 RALGPS2Ral GEF with PH domain and SH3 binding motif 2 (220338_at), score: -0.87 RICH2Rho-type GTPase-activating protein RICH2 (215232_at), score: -0.88 RPS4Y1ribosomal protein S4, Y-linked 1 (201909_at), score: 0.62 S100A14S100 calcium binding protein A14 (218677_at), score: -0.88 SCARB1scavenger receptor class B, member 1 (201819_at), score: 0.66 SCG2secretogranin II (chromogranin C) (204035_at), score: 0.7 SETXsenataxin (201964_at), score: 0.64 SIN3BSIN3 homolog B, transcription regulator (yeast) (209352_s_at), score: 0.77 SLC1A7solute carrier family 1 (glutamate transporter), member 7 (210923_at), score: -0.79 SLC26A10solute carrier family 26, member 10 (214951_at), score: -0.87 SLC9A1solute carrier family 9 (sodium/hydrogen exchanger), member 1 (209453_at), score: 0.68 SNORA21small nucleolar RNA, H/ACA box 21 (215224_at), score: -0.8 SPARCL1SPARC-like 1 (hevin) (200795_at), score: -0.83 SPRY4sprouty homolog 4 (Drosophila) (221489_s_at), score: 0.62 TBX21T-box 21 (220684_at), score: -0.91 TNFSF4tumor necrosis factor (ligand) superfamily, member 4 (207426_s_at), score: 0.69 VEGFBvascular endothelial growth factor B (203683_s_at), score: 0.7 VPS37Bvacuolar protein sorting 37 homolog B (S. cerevisiae) (221704_s_at), score: 0.67 ZDHHC11zinc finger, DHHC-type containing 11 (221646_s_at), score: -0.88 ZNF282zinc finger protein 282 (212892_at), score: 0.61 ZPBPzona pellucida binding protein (207021_at), score: -0.83
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-3407-raw-cel-1437949557.cel | 1 | 4 | Cockayne | hgu133a | CS | eGFP |
| 2Twin.CEL | 2 | 2 | DS-twin | hgu133plus2 | none | DS-twin 2 |
| 6Twin.CEL | 6 | 2 | DS-twin | hgu133plus2 | none | DS-twin 6 |
| t21d 08-03.CEL | 7 | 1 | DS-CC | hgu133a | Down | DS-CC 7 |
| t21a 08-03.CEL | 4 | 1 | DS-CC | hgu133a | Down | DS-CC 4 |
| E-TABM-263-raw-cel-1515486411.cel | 39 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486031.cel | 20 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 46A.CEL | 1 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 1 |
| E-TABM-263-raw-cel-1515486071.cel | 22 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |