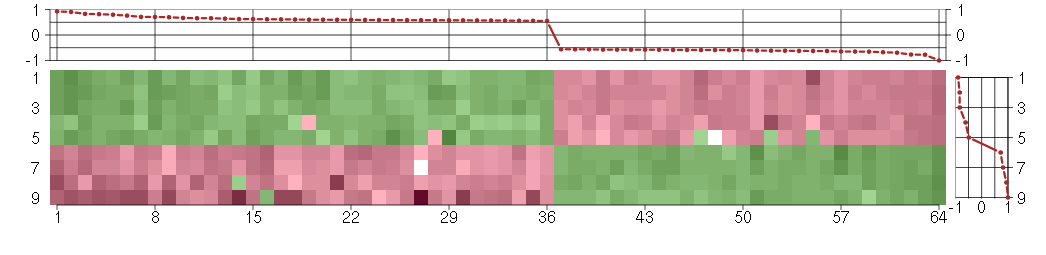

Under-expression is coded with green,
over-expression with red color.

multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
nervous system development
The process whose specific outcome is the progression of nervous tissue over time, from its formation to its mature state.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
neurogenesis
Generation of cells within the nervous system.
cell differentiation
The process whereby relatively unspecialized cells, e.g. embryonic or regenerative cells, acquire specialized structural and/or functional features that characterize the cells, tissues, or organs of the mature organism or some other relatively stable phase of the organism's life history. Differentiation includes the processes involved in commitment of a cell to a specific fate.
neuron differentiation
The process whereby a relatively unspecialized cell acquires specialized features of a neuron.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
developmental process
A biological process whose specific outcome is the progression of an integrated living unit: an anatomical structure (which may be a subcellular structure, cell, tissue, or organ), or organism over time from an initial condition to a later condition.
regulation of cell differentiation
Any process that modulates the frequency, rate or extent of cell differentiation, the process whereby relatively unspecialized cells acquire specialized structural and functional features.
negative regulation of cell differentiation
Any process that stops, prevents or reduces the frequency, rate or extent of cell differentiation.
regulation of neuron differentiation
Any process that modulates the frequency, rate or extent of neuron differentiation.
negative regulation of neuron differentiation
Any process that stops, prevents or reduces the frequency, rate or extent of neuron differentiation.
cell development
The process whose specific outcome is the progression of the cell over time, from its formation to the mature structure. Cell development does not include the steps involved in committing a cell to a specific fate.
negative regulation of biological process
Any process that stops, prevents or reduces the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
negative regulation of cellular process
Any process that stops, prevents or reduces the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
generation of neurons
The process by which nerve cells are generated. This includes the production of neuroblasts and their differentiation into neurons.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
anatomical structure development
The biological process whose specific outcome is the progression of an anatomical structure from an initial condition to its mature state. This process begins with the formation of the structure and ends with the mature structure, whatever form that may be including its natural destruction. An anatomical structure is any biological entity that occupies space and is distinguished from its surroundings. Anatomical structures can be macroscopic such as a carpel, or microscopic such as an acrosome.
cellular developmental process
A biological process whose specific outcome is the progression of a cell over time from an initial condition to a later condition.
regulation of neurogenesis
Any process that modulates the frequency, rate or extent of neurogenesis, the origin and formation of neurons.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of developmental process
Any process that modulates the frequency, rate or extent of development, the biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote, or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
negative regulation of developmental process
Any process that stops, prevents or reduces the rate or extent of development, the biological process whose specific outcome is the progression of an organism over time from an initial condition (e.g. a zygote, or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
regulation of multicellular organismal process
Any process that modulates the frequency, rate or extent of a multicellular organismal process, the processes pertinent to the function of a multicellular organism above the cellular level; includes the integrated processes of tissues and organs.
regulation of nervous system development
Any process that modulates the frequency, rate or extent of nervous system development, the origin and formation of nervous tissue.
regulation of cell development
Any process that modulates the rate, frequency or extent of the progression of the cell over time, from its formation to the mature structure. Cell development does not include the steps involved in committing a cell to a specific fate.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
all
This term is the most general term possible
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
cellular developmental process
A biological process whose specific outcome is the progression of a cell over time from an initial condition to a later condition.
negative regulation of cellular process
Any process that stops, prevents or reduces the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
negative regulation of developmental process
Any process that stops, prevents or reduces the rate or extent of development, the biological process whose specific outcome is the progression of an organism over time from an initial condition (e.g. a zygote, or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
negative regulation of biological process
Any process that stops, prevents or reduces the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of developmental process
Any process that modulates the frequency, rate or extent of development, the biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote, or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of multicellular organismal process
Any process that modulates the frequency, rate or extent of a multicellular organismal process, the processes pertinent to the function of a multicellular organism above the cellular level; includes the integrated processes of tissues and organs.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
negative regulation of cellular process
Any process that stops, prevents or reduces the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
regulation of cell differentiation
Any process that modulates the frequency, rate or extent of cell differentiation, the process whereby relatively unspecialized cells acquire specialized structural and functional features.
negative regulation of developmental process
Any process that stops, prevents or reduces the rate or extent of development, the biological process whose specific outcome is the progression of an organism over time from an initial condition (e.g. a zygote, or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
regulation of nervous system development
Any process that modulates the frequency, rate or extent of nervous system development, the origin and formation of nervous tissue.
negative regulation of cell differentiation
Any process that stops, prevents or reduces the frequency, rate or extent of cell differentiation.
regulation of neurogenesis
Any process that modulates the frequency, rate or extent of neurogenesis, the origin and formation of neurons.
regulation of cell differentiation
Any process that modulates the frequency, rate or extent of cell differentiation, the process whereby relatively unspecialized cells acquire specialized structural and functional features.
negative regulation of cell differentiation
Any process that stops, prevents or reduces the frequency, rate or extent of cell differentiation.
cell development
The process whose specific outcome is the progression of the cell over time, from its formation to the mature structure. Cell development does not include the steps involved in committing a cell to a specific fate.
negative regulation of cell differentiation
Any process that stops, prevents or reduces the frequency, rate or extent of cell differentiation.
regulation of cell development
Any process that modulates the rate, frequency or extent of the progression of the cell over time, from its formation to the mature structure. Cell development does not include the steps involved in committing a cell to a specific fate.
regulation of neurogenesis
Any process that modulates the frequency, rate or extent of neurogenesis, the origin and formation of neurons.
neuron differentiation
The process whereby a relatively unspecialized cell acquires specialized features of a neuron.
regulation of neurogenesis
Any process that modulates the frequency, rate or extent of neurogenesis, the origin and formation of neurons.
regulation of neuron differentiation
Any process that modulates the frequency, rate or extent of neuron differentiation.
negative regulation of neuron differentiation
Any process that stops, prevents or reduces the frequency, rate or extent of neuron differentiation.
neurogenesis
Generation of cells within the nervous system.
regulation of nervous system development
Any process that modulates the frequency, rate or extent of nervous system development, the origin and formation of nervous tissue.
negative regulation of neuron differentiation
Any process that stops, prevents or reduces the frequency, rate or extent of neuron differentiation.


molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
catalytic activity
Catalysis of a biochemical reaction at physiological temperatures. In biologically catalyzed reactions, the reactants are known as substrates, and the catalysts are naturally occurring macromolecular substances known as enzymes. Enzymes possess specific binding sites for substrates, and are usually composed wholly or largely of protein, but RNA that has catalytic activity (ribozyme) is often also regarded as enzymatic.
aldo-keto reductase activity
Catalysis of the NADPH-dependent reduction of carbonyl compounds.
oxidoreductase activity
Catalysis of an oxidation-reduction (redox) reaction, a reversible chemical reaction in which the oxidation state of an atom or atoms within a molecule is altered. One substrate acts as a hydrogen or electron donor and becomes oxidized, while the other acts as hydrogen or electron acceptor and becomes reduced.
steroid dehydrogenase activity
Catalysis of an oxidation-reduction (redox) reaction in which one substrate is a sterol derivative.
oxidoreductase activity, acting on CH-OH group of donors
Catalysis of an oxidation-reduction (redox) reaction in which a CH-OH group act as a hydrogen or electron donor and reduces a hydrogen or electron acceptor.
oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor
Catalysis of an oxidation-reduction (redox) reaction in which a CH-OH group acts as a hydrogen or electron donor and reduces NAD+ or NADP.
steroid dehydrogenase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor
Catalysis of an oxidation-reduction (redox) reaction in which a CH-OH group acts as a hydrogen or electron donor and reduces NAD+ or NADP, and in which one substrate is a sterol derivative.
3-alpha-hydroxysteroid dehydrogenase (A-specific) activity
Catalysis of the reaction: NAD(P)+ + androsterone = NAD(P)H + H+ + 5-alpha-androstane-3,17-dione. The reaction is A-specific (i.e. the pro-R hydrogen is transferred from the 4-position of reduced nicotinamide cofactor) with respect to NAD(P)+.
all
This term is the most general term possible
steroid dehydrogenase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor
Catalysis of an oxidation-reduction (redox) reaction in which a CH-OH group acts as a hydrogen or electron donor and reduces NAD+ or NADP, and in which one substrate is a sterol derivative.
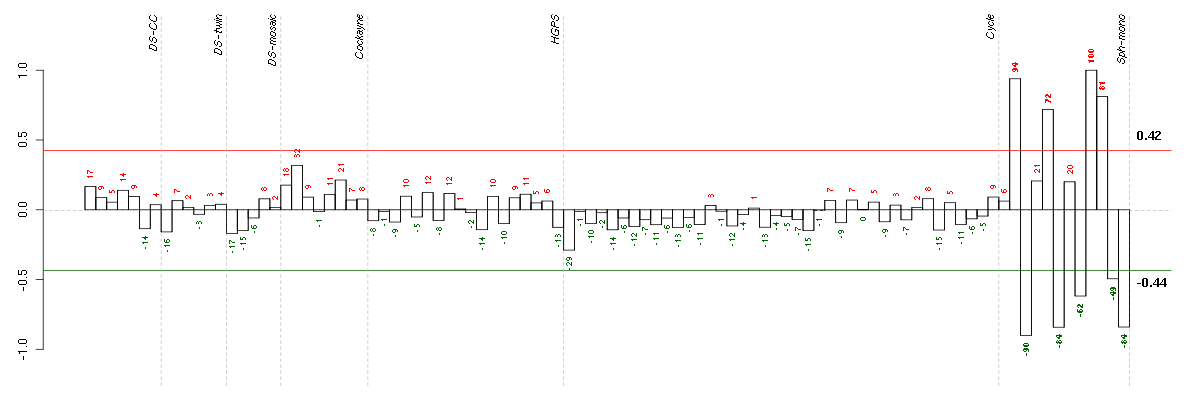
ACSL5acyl-CoA synthetase long-chain family member 5 (218322_s_at), score: 0.58 AKR1B10aldo-keto reductase family 1, member B10 (aldose reductase) (206561_s_at), score: 0.82 AKR1C2aldo-keto reductase family 1, member C2 (dihydrodiol dehydrogenase 2; bile acid binding protein; 3-alpha hydroxysteroid dehydrogenase, type III) (209699_x_at), score: 0.57 AKR1C3aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II) (209160_at), score: 0.57 ALDH3A1aldehyde dehydrogenase 3 family, memberA1 (205623_at), score: 0.62 ALDH3A2aldehyde dehydrogenase 3 family, member A2 (202053_s_at), score: 0.58 ARMCX1armadillo repeat containing, X-linked 1 (218694_at), score: -0.58 ASB9ankyrin repeat and SOCS box-containing 9 (205673_s_at), score: 0.62 BGNbiglycan (213905_x_at), score: -0.62 BMP7bone morphogenetic protein 7 (209590_at), score: 0.61 C14orf169chromosome 14 open reading frame 169 (219526_at), score: -0.69 C20orf39chromosome 20 open reading frame 39 (219310_at), score: 0.71 CAND2cullin-associated and neddylation-dissociated 2 (putative) (213547_at), score: -0.57 CD24CD24 molecule (266_s_at), score: 0.59 CELSR1cadherin, EGF LAG seven-pass G-type receptor 1 (flamingo homolog, Drosophila) (41660_at), score: 0.79 CELSR3cadherin, EGF LAG seven-pass G-type receptor 3 (flamingo homolog, Drosophila) (40020_at), score: 0.71 CLEC11AC-type lectin domain family 11, member A (211709_s_at), score: -0.56 COL3A1collagen, type III, alpha 1 (215076_s_at), score: -0.63 CUGBP2CUG triplet repeat, RNA binding protein 2 (202158_s_at), score: -0.56 DGKDdiacylglycerol kinase, delta 130kDa (208072_s_at), score: 0.6 DLX4distal-less homeobox 4 (208216_at), score: 0.58 EGLN3egl nine homolog 3 (C. elegans) (219232_s_at), score: 0.55 EIF4A1eukaryotic translation initiation factor 4A, isoform 1 (211787_s_at), score: 0.59 EMP1epithelial membrane protein 1 (201325_s_at), score: -0.61 FAM174Bfamily with sequence similarity 174, member B (51158_at), score: -0.63 FCGR2AFc fragment of IgG, low affinity IIa, receptor (CD32) (203561_at), score: 0.7 FGF13fibroblast growth factor 13 (205110_s_at), score: 0.57 FOXG1forkhead box G1 (206018_at), score: 0.55 GABRA2gamma-aminobutyric acid (GABA) A receptor, alpha 2 (207014_at), score: 0.58 GALCgalactosylceramidase (204417_at), score: -0.59 GREM1gremlin 1, cysteine knot superfamily, homolog (Xenopus laevis) (218469_at), score: -1 IGSF3immunoglobulin superfamily, member 3 (202421_at), score: 0.83 ISL1ISL LIM homeobox 1 (206104_at), score: -0.59 KYNUkynureninase (L-kynurenine hydrolase) (217388_s_at), score: 0.9 LAPTM5lysosomal multispanning membrane protein 5 (201721_s_at), score: 0.59 LEF1lymphoid enhancer-binding factor 1 (221558_s_at), score: 0.76 LUMlumican (201744_s_at), score: -0.77 MED31mediator complex subunit 31 (219318_x_at), score: 0.56 MPPED2metallophosphoesterase domain containing 2 (205413_at), score: 0.93 MYO1Bmyosin IB (212364_at), score: -0.77 NPAS2neuronal PAS domain protein 2 (39549_at), score: -0.58 NPTX1neuronal pentraxin I (204684_at), score: 0.6 PCLOpiccolo (presynaptic cytomatrix protein) (213558_at), score: 0.62 PDGFRAplatelet-derived growth factor receptor, alpha polypeptide (203131_at), score: -0.66 PSTPIP2proline-serine-threonine phosphatase interacting protein 2 (219938_s_at), score: -0.65 PTPRDprotein tyrosine phosphatase, receptor type, D (214043_at), score: -0.62 RARBretinoic acid receptor, beta (205080_at), score: 0.64 RIPK4receptor-interacting serine-threonine kinase 4 (221215_s_at), score: 0.67 RND3Rho family GTPase 3 (212724_at), score: -0.65 SEMA3Fsema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F (35666_at), score: -0.58 SERPINB9serpin peptidase inhibitor, clade B (ovalbumin), member 9 (209723_at), score: -0.59 SH3BGRSH3 domain binding glutamic acid-rich protein (204979_s_at), score: 0.6 SLC30A1solute carrier family 30 (zinc transporter), member 1 (212907_at), score: -0.56 SPANXCSPANX family, member C (220217_x_at), score: 0.66 TCF7L2transcription factor 7-like 2 (T-cell specific, HMG-box) (212761_at), score: -0.63 TEStestis derived transcript (3 LIM domains) (202720_at), score: -0.67 TGFB2transforming growth factor, beta 2 (220407_s_at), score: 0.66 TGFBItransforming growth factor, beta-induced, 68kDa (201506_at), score: 0.58 TGIF2TGFB-induced factor homeobox 2 (216262_s_at), score: 0.6 THBS2thrombospondin 2 (203083_at), score: -0.59 TNS1tensin 1 (221748_s_at), score: -0.58 TPBGtrophoblast glycoprotein (203476_at), score: -0.59 TSPYL5TSPY-like 5 (213122_at), score: -0.59 VAV3vav 3 guanine nucleotide exchange factor (218807_at), score: 0.63
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-4219-raw-cel-1311956178.cel | 6 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956358.cel | 10 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956824.cel | 24 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956418.cel | 13 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956634.cel | 19 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956321.cel | 9 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956614.cel | 18 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956138.cel | 4 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956457.cel | 14 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |