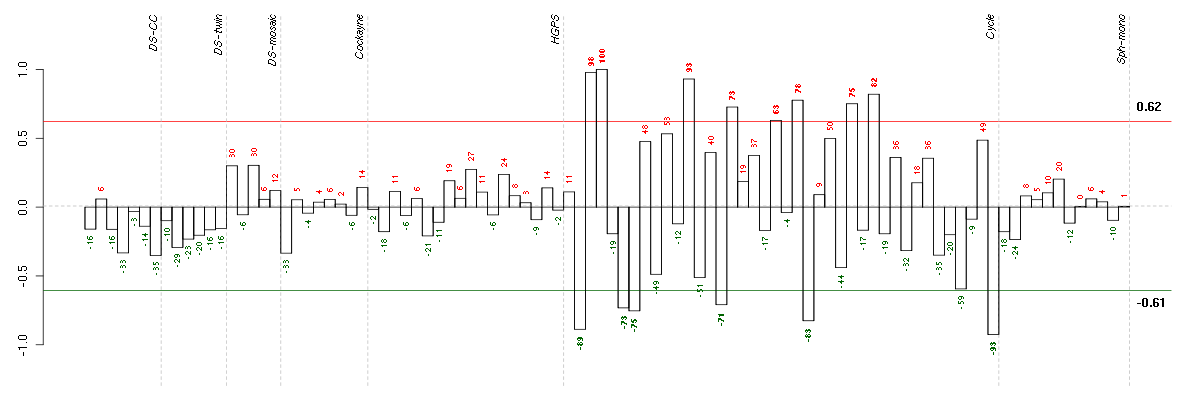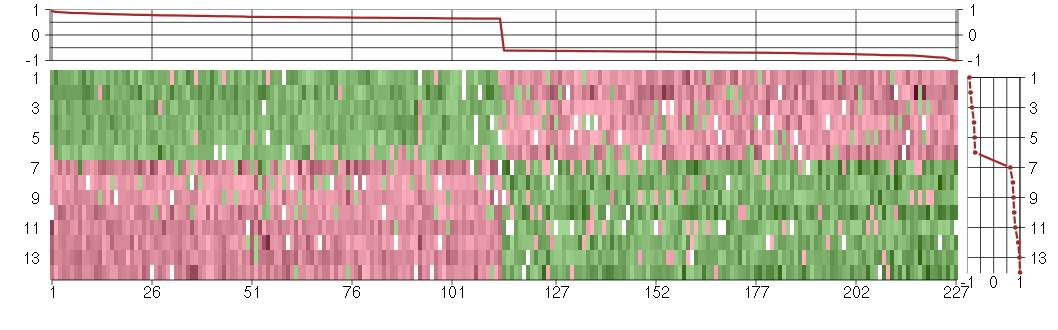

Under-expression is coded with green,
over-expression with red color.



ABCA7ATP-binding cassette, sub-family A (ABC1), member 7 (219577_s_at), score: -0.81 ABCG4ATP-binding cassette, sub-family G (WHITE), member 4 (207593_at), score: -0.62 ABLIM1actin binding LIM protein 1 (200965_s_at), score: 0.83 ACTN3actinin, alpha 3 (206891_at), score: -0.79 ADAadenosine deaminase (204639_at), score: -0.65 ADCK2aarF domain containing kinase 2 (221893_s_at), score: 0.83 ADCY3adenylate cyclase 3 (209321_s_at), score: 0.66 ADORA1adenosine A1 receptor (216220_s_at), score: -0.64 AGPAT11-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) (215535_s_at), score: 0.69 ALDH1L1aldehyde dehydrogenase 1 family, member L1 (215798_at), score: -0.8 ALPPL2alkaline phosphatase, placental-like 2 (216377_x_at), score: -0.61 APOC4apolipoprotein C-IV (206738_at), score: -0.71 ARHGAP22Rho GTPase activating protein 22 (206298_at), score: 0.81 ARL17P1ADP-ribosylation factor-like 17 pseudogene 1 (210718_s_at), score: 0.69 ATF5activating transcription factor 5 (204999_s_at), score: 0.69 ATMataxia telangiectasia mutated (210858_x_at), score: -0.77 B3GAT3beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) (203452_at), score: 0.67 BAP1BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) (201419_at), score: 0.69 BAT2HLA-B associated transcript 2 (212081_x_at), score: 0.81 BAT2LHLA-B associated transcript 2-like (212068_s_at), score: 0.68 BTBD2BTB (POZ) domain containing 2 (207722_s_at), score: 0.7 BTNL3butyrophilin-like 3 (217207_s_at), score: -0.7 C16orf57chromosome 16 open reading frame 57 (218060_s_at), score: 0.68 C19orf61chromosome 19 open reading frame 61 (221335_x_at), score: 0.84 C1orf38chromosome 1 open reading frame 38 (210785_s_at), score: 0.67 C1orf69chromosome 1 open reading frame 69 (215490_at), score: -0.63 C20orf111chromosome 20 open reading frame 111 (209020_at), score: -0.7 C2orf37chromosome 2 open reading frame 37 (220172_at), score: 0.71 C7orf58chromosome 7 open reading frame 58 (220032_at), score: 0.64 CALB2calbindin 2 (205428_s_at), score: -0.72 CASP7caspase 7, apoptosis-related cysteine peptidase (207181_s_at), score: -0.65 CD79BCD79b molecule, immunoglobulin-associated beta (205297_s_at), score: -1 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (217849_s_at), score: 0.88 CDC42EP4CDC42 effector protein (Rho GTPase binding) 4 (218062_x_at), score: 0.73 CICcapicua homolog (Drosophila) (212784_at), score: 0.82 CITcitron (rho-interacting, serine/threonine kinase 21) (212801_at), score: 0.68 CIZ1CDKN1A interacting zinc finger protein 1 (205516_x_at), score: 0.67 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: 0.84 COL11A2collagen, type XI, alpha 2 (216993_s_at), score: -0.79 COL18A1collagen, type XVIII, alpha 1 (209081_s_at), score: 0.64 CORO2Bcoronin, actin binding protein, 2B (209789_at), score: 0.75 COX6A2cytochrome c oxidase subunit VIa polypeptide 2 (206353_at), score: -0.68 CYP3A4cytochrome P450, family 3, subfamily A, polypeptide 4 (205998_x_at), score: -0.82 CYTH4cytohesin 4 (219183_s_at), score: -0.77 DENND3DENN/MADD domain containing 3 (212974_at), score: 0.7 DENND4BDENN/MADD domain containing 4B (202860_at), score: 0.71 DLSTPdihydrolipoamide S-succinyltransferase pseudogene (E2 component of 2-oxo-glutarate complex) (215210_s_at), score: -0.69 DLX4distal-less homeobox 4 (208216_at), score: -0.62 DMBT1deleted in malignant brain tumors 1 (208250_s_at), score: -0.63 DMPKdystrophia myotonica-protein kinase (37996_s_at), score: 0.64 DSPPdentin sialophosphoprotein (221681_s_at), score: -0.79 DUSP8dual specificity phosphatase 8 (206374_at), score: -0.63 EDARectodysplasin A receptor (220048_at), score: -0.67 EHBP1L1EH domain binding protein 1-like 1 (221755_at), score: 0.65 EMCNendomucin (219436_s_at), score: 0.71 EPHA4EPH receptor A4 (206114_at), score: -0.71 EPM2Aepilepsy, progressive myoclonus type 2A, Lafora disease (laforin) (205231_s_at), score: -0.71 EVLEnah/Vasp-like (217838_s_at), score: 0.73 F11coagulation factor XI (206610_s_at), score: -0.79 FBXO2F-box protein 2 (219305_x_at), score: -0.75 FCER1GFc fragment of IgE, high affinity I, receptor for; gamma polypeptide (204232_at), score: -0.62 FGL1fibrinogen-like 1 (205305_at), score: -0.7 FKBP10FK506 binding protein 10, 65 kDa (219249_s_at), score: -0.66 FKRPfukutin related protein (219853_at), score: 0.73 FLRT3fibronectin leucine rich transmembrane protein 3 (219250_s_at), score: 0.75 FMO3flavin containing monooxygenase 3 (40665_at), score: 0.65 FNDC4fibronectin type III domain containing 4 (218843_at), score: 0.65 FOXC2forkhead box C2 (MFH-1, mesenchyme forkhead 1) (214520_at), score: 0.67 FOXK2forkhead box K2 (203064_s_at), score: 0.91 GABARAPL1GABA(A) receptor-associated protein like 1 (208868_s_at), score: -0.69 GABARAPL3GABA(A) receptors associated protein like 3 (pseudogene) (211458_s_at), score: -0.73 GBA3glucosidase, beta, acid 3 (cytosolic) (219954_s_at), score: -0.79 GDF9growth differentiation factor 9 (221314_at), score: -0.64 GGA3golgi associated, gamma adaptin ear containing, ARF binding protein 3 (209411_s_at), score: 0.79 GNL3Lguanine nucleotide binding protein-like 3 (nucleolar)-like (205010_at), score: -0.73 GNL3LPguanine nucleotide binding protein-like 3 (nucleolar)-like pseudogene (220716_at), score: -0.68 GOLGA8Ggolgi autoantigen, golgin subfamily a, 8G (222149_x_at), score: -0.69 GPR144G protein-coupled receptor 144 (216289_at), score: -0.88 GPR56G protein-coupled receptor 56 (212070_at), score: 0.77 GTF2H3general transcription factor IIH, polypeptide 3, 34kDa (222104_x_at), score: -0.66 HAB1B1 for mucin (215778_x_at), score: -0.93 HAMPhepcidin antimicrobial peptide (220491_at), score: -0.7 HGFhepatocyte growth factor (hepapoietin A; scatter factor) (209960_at), score: 0.67 HLA-DQB1major histocompatibility complex, class II, DQ beta 1 (211654_x_at), score: -0.64 HMGA1high mobility group AT-hook 1 (210457_x_at), score: 0.76 HNRNPH2heterogeneous nuclear ribonucleoprotein H2 (H') (201132_at), score: -0.65 HOXB5homeobox B5 (205601_s_at), score: -0.61 HSD17B6hydroxysteroid (17-beta) dehydrogenase 6 homolog (mouse) (37512_at), score: -0.69 IDUAiduronidase, alpha-L- (205059_s_at), score: 0.9 IGHG1immunoglobulin heavy constant gamma 1 (G1m marker) (211693_at), score: -0.83 IL9Rinterleukin 9 receptor (217212_s_at), score: -0.86 INAinternexin neuronal intermediate filament protein, alpha (204465_s_at), score: 0.7 INE1inactivation escape 1 (non-protein coding) (207252_at), score: -0.65 INTS1integrator complex subunit 1 (212212_s_at), score: 0.65 IRF2interferon regulatory factor 2 (203275_at), score: 0.66 ITGALintegrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide) (213475_s_at), score: -0.77 JAG2jagged 2 (32137_at), score: -0.63 JUPjunction plakoglobin (201015_s_at), score: 0.8 KATNB1katanin p80 (WD repeat containing) subunit B 1 (203162_s_at), score: -0.62 KCNJ14potassium inwardly-rectifying channel, subfamily J, member 14 (220776_at), score: -0.61 KCTD15potassium channel tetramerisation domain containing 15 (218553_s_at), score: -0.73 KCTD2potassium channel tetramerisation domain containing 2 (212564_at), score: 0.66 KIAA0892KIAA0892 (212505_s_at), score: 0.69 KIAA1009KIAA1009 (206005_s_at), score: -0.63 KIAA1305KIAA1305 (220911_s_at), score: 0.75 KRT24keratin 24 (220267_at), score: -0.67 KRT8keratin 8 (209008_x_at), score: -0.62 LARSleucyl-tRNA synthetase (217810_x_at), score: -0.68 LEPREL2leprecan-like 2 (204854_at), score: 0.86 LILRB3leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3 (211133_x_at), score: -0.69 LLGL2lethal giant larvae homolog 2 (Drosophila) (203713_s_at), score: -0.76 LOC100129500hypothetical protein LOC100129500 (212884_x_at), score: -0.64 LOC100133105hypothetical protein LOC100133105 (214237_x_at), score: -0.67 LOC128192similar to peptidyl-Pro cis trans isomerase (217346_at), score: -0.69 LOC149478hypothetical protein LOC149478 (215462_at), score: -0.64 LOC149501similar to keratin 8 (216821_at), score: -0.65 LOC391132similar to hCG2041276 (216177_at), score: -0.69 LOC80054hypothetical LOC80054 (220465_at), score: -0.71 LOC81691exonuclease NEF-sp (208107_s_at), score: 0.7 LPHN1latrophilin 1 (203488_at), score: -0.64 LRDDleucine-rich repeats and death domain containing (221640_s_at), score: 0.83 MAFKv-maf musculoaponeurotic fibrosarcoma oncogene homolog K (avian) (206750_at), score: -0.61 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: 0.74 MAP3K6mitogen-activated protein kinase kinase kinase 6 (219278_at), score: 0.79 MAPKAPK3mitogen-activated protein kinase-activated protein kinase 3 (202788_at), score: 0.77 MAST4microtubule associated serine/threonine kinase family member 4 (40016_g_at), score: 0.78 MED15mediator complex subunit 15 (222175_s_at), score: 0.67 MEOX1mesenchyme homeobox 1 (205619_s_at), score: -0.62 MGAT1mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase (201126_s_at), score: 0.68 MICAL3microtubule associated monoxygenase, calponin and LIM domain containing 3 (212715_s_at), score: 0.71 MINK1misshapen-like kinase 1 (zebrafish) (214246_x_at), score: 0.68 MOCS1molybdenum cofactor synthesis 1 (211673_s_at), score: -0.72 MRPS6mitochondrial ribosomal protein S6 (213167_s_at), score: 0.69 MTMR11myotubularin related protein 11 (205076_s_at), score: 0.73 MUC5ACmucin 5AC, oligomeric mucus/gel-forming (217182_at), score: -0.75 MUL1mitochondrial E3 ubiquitin ligase 1 (218246_at), score: 0.79 MYL4myosin, light chain 4, alkali; atrial, embryonic (210088_x_at), score: -0.69 NBL1neuroblastoma, suppression of tumorigenicity 1 (37005_at), score: 0.65 NCRNA00093non-protein coding RNA 93 (210723_x_at), score: -0.61 NDPNorrie disease (pseudoglioma) (206022_at), score: 0.76 NF1neurofibromin 1 (211094_s_at), score: 0.71 NFATC4nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 (205897_at), score: 0.71 NFYCnuclear transcription factor Y, gamma (202215_s_at), score: 0.69 NOVA2neuro-oncological ventral antigen 2 (206477_s_at), score: -0.74 NUPL2nucleoporin like 2 (204003_s_at), score: -0.7 NXPH3neurexophilin 3 (221991_at), score: -0.73 OAS32'-5'-oligoadenylate synthetase 3, 100kDa (218400_at), score: 0.77 OLFML2Bolfactomedin-like 2B (213125_at), score: 0.94 OLR1oxidized low density lipoprotein (lectin-like) receptor 1 (210004_at), score: -0.65 OPRL1opiate receptor-like 1 (206564_at), score: -0.74 OR7E47Polfactory receptor, family 7, subfamily E, member 47 pseudogene (216698_x_at), score: -0.65 PAK1p21 protein (Cdc42/Rac)-activated kinase 1 (209615_s_at), score: 0.71 PARVBparvin, beta (37965_at), score: 0.86 PHF7PHD finger protein 7 (215622_x_at), score: -0.7 PIGOphosphatidylinositol glycan anchor biosynthesis, class O (209998_at), score: 0.66 PIP5K1Cphosphatidylinositol-4-phosphate 5-kinase, type I, gamma (212518_at), score: 0.76 PKLRpyruvate kinase, liver and RBC (222078_at), score: -0.73 PKN1protein kinase N1 (202161_at), score: 0.67 POLR2Apolymerase (RNA) II (DNA directed) polypeptide A, 220kDa (202725_at), score: 0.69 POLRMTpolymerase (RNA) mitochondrial (DNA directed) (203782_s_at), score: 0.64 POM121POM121 membrane glycoprotein (rat) (212178_s_at), score: 0.7 POM121L2POM121 membrane glycoprotein-like 2 (rat) (216582_at), score: -0.66 PPP1R13Bprotein phosphatase 1, regulatory (inhibitor) subunit 13B (216347_s_at), score: -0.64 PRKCDprotein kinase C, delta (202545_at), score: 0.73 PRR14proline rich 14 (218714_at), score: 0.7 PTPN6protein tyrosine phosphatase, non-receptor type 6 (206687_s_at), score: -0.85 PXNpaxillin (211823_s_at), score: 0.86 RASGRP3RAS guanyl releasing protein 3 (calcium and DAG-regulated) (205801_s_at), score: 0.66 RBM3RNA binding motif (RNP1, RRM) protein 3 (222026_at), score: -0.63 RBM38RNA binding motif protein 38 (212430_at), score: -0.66 REREarginine-glutamic acid dipeptide (RE) repeats (200940_s_at), score: 0.65 RLN1relaxin 1 (211753_s_at), score: -0.62 RNF220ring finger protein 220 (219988_s_at), score: 0.79 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: -0.99 RPRD2regulation of nuclear pre-mRNA domain containing 2 (212553_at), score: 0.7 RPS17P5ribosomal protein S17 pseudogene 5 (216348_at), score: -0.8 RSAD1radical S-adenosyl methionine domain containing 1 (218307_at), score: 0.78 SAA4serum amyloid A4, constitutive (207096_at), score: -0.72 SBNO2strawberry notch homolog 2 (Drosophila) (204166_at), score: 0.68 SCAMP4secretory carrier membrane protein 4 (213244_at), score: 0.85 SEMA3Fsema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F (35666_at), score: -0.68 SEPT5septin 5 (209767_s_at), score: -0.63 SF1splicing factor 1 (208313_s_at), score: 0.75 SH2B3SH2B adaptor protein 3 (203320_at), score: 0.64 SH2D3ASH2 domain containing 3A (222169_x_at), score: -0.65 SIGLEC1sialic acid binding Ig-like lectin 1, sialoadhesin (44673_at), score: -0.75 SIN3BSIN3 homolog B, transcription regulator (yeast) (209352_s_at), score: 0.7 SIRT6sirtuin (silent mating type information regulation 2 homolog) 6 (S. cerevisiae) (219613_s_at), score: 0.86 SLC26A10solute carrier family 26, member 10 (214951_at), score: -0.61 SLC29A1solute carrier family 29 (nucleoside transporters), member 1 (201801_s_at), score: 0.64 SLC5A5solute carrier family 5 (sodium iodide symporter), member 5 (211123_at), score: -0.76 SMG7Smg-7 homolog, nonsense mediated mRNA decay factor (C. elegans) (201794_s_at), score: 0.74 SMYD5SMYD family member 5 (209516_at), score: 0.66 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: 0.81 SPATA1spermatogenesis associated 1 (221057_at), score: -0.67 SPG7spastic paraplegia 7 (pure and complicated autosomal recessive) (202104_s_at), score: 0.73 ST8SIA3ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 (208064_s_at), score: -0.68 STIM1stromal interaction molecule 1 (202764_at), score: 0.77 SULT1B1sulfotransferase family, cytosolic, 1B, member 1 (207601_at), score: -0.64 TAF15TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa (202840_at), score: -0.63 TAPBPTAP binding protein (tapasin) (208829_at), score: 0.69 TENC1tensin like C1 domain containing phosphatase (tensin 2) (212494_at), score: 0.65 TIAF1TGFB1-induced anti-apoptotic factor 1 (202039_at), score: 0.7 TICAM2toll-like receptor adaptor molecule 2 (214658_at), score: -0.66 TLE6transducin-like enhancer of split 6 (E(sp1) homolog, Drosophila) (222219_s_at), score: -0.72 TMCC2transmembrane and coiled-coil domain family 2 (213096_at), score: -0.69 TMSB4Ythymosin beta 4, Y-linked (206769_at), score: -0.89 TRMT2ATRM2 tRNA methyltransferase 2 homolog A (S. cerevisiae) (91617_at), score: -0.65 TSC2tuberous sclerosis 2 (215735_s_at), score: 0.65 TULP2tubby like protein 2 (206733_at), score: -0.8 TXNthioredoxin (216609_at), score: -0.63 UBTD1ubiquitin domain containing 1 (219172_at), score: 0.76 ULK1unc-51-like kinase 1 (C. elegans) (209333_at), score: 0.66 UPF1UPF1 regulator of nonsense transcripts homolog (yeast) (211168_s_at), score: 0.72 VCPIP1valosin containing protein (p97)/p47 complex interacting protein 1 (219810_at), score: -0.63 VEGFBvascular endothelial growth factor B (203683_s_at), score: 0.89 VPS37Bvacuolar protein sorting 37 homolog B (S. cerevisiae) (221704_s_at), score: 0.8 WDR42AWD repeat domain 42A (202249_s_at), score: 0.7 WDR6WD repeat domain 6 (217734_s_at), score: 0.67 WNT2Bwingless-type MMTV integration site family, member 2B (206459_s_at), score: -0.68 XAB2XPA binding protein 2 (218110_at), score: 0.68 YES1v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 (202932_at), score: 0.67 YPEL1yippee-like 1 (Drosophila) (206063_x_at), score: -0.79 ZNF132zinc finger protein 132 (207402_at), score: 0.64 ZNF281zinc finger protein 281 (218401_s_at), score: -0.73 ZNF318zinc finger protein 318 (203521_s_at), score: 0.77
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485931.cel | 15 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486031.cel | 20 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485951.cel | 16 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486171.cel | 27 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486071.cel | 22 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |