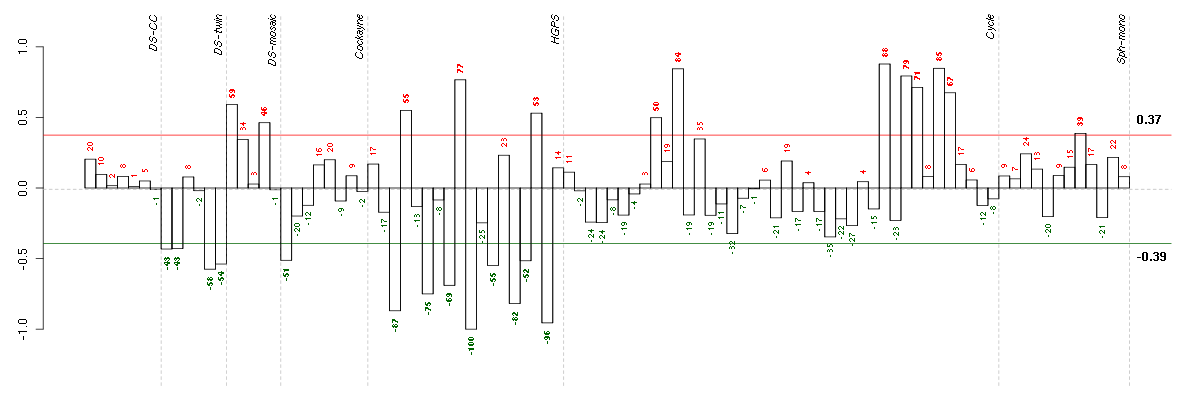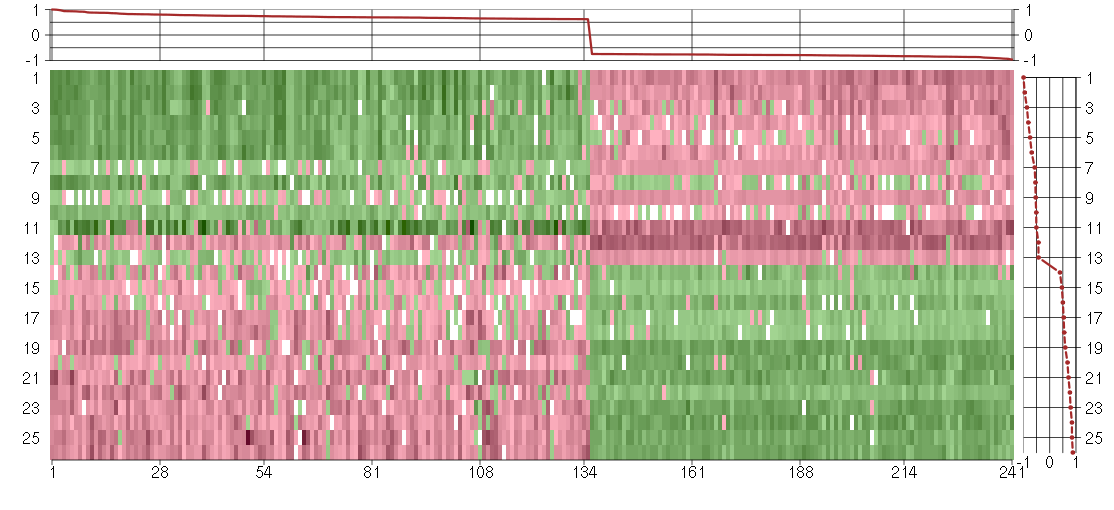

Under-expression is coded with green,
over-expression with red color.

DNA replication
The process whereby new strands of DNA are synthesized. The template for replication can either be an existing DNA molecule or RNA.
cell cycle checkpoint
The cell cycle regulatory process by which progression through the cycle can be halted until conditions are suitable for the cell to proceed to the next stage.
M phase of mitotic cell cycle
A cell cycle process comprising the steps by which a cell progresses through M phase, the part of the mitotic cell cycle during which mitosis takes place.
mitotic cell cycle
Progression through the phases of the mitotic cell cycle, the most common eukaryotic cell cycle, which canonically comprises four successive phases called G1, S, G2, and M and includes replication of the genome and the subsequent segregation of chromosomes into daughter cells. In some variant cell cycles nuclear replication or nuclear division may not be followed by cell division, or G1 and G2 phases may be absent.
M phase
A cell cycle process comprising the steps by which a cell progresses through M phase, the part of the cell cycle comprising nuclear division.
nuclear division
A process by which a cell nucleus is divided into two nuclei, with DNA and other nuclear contents distributed between the daughter nuclei.
DNA synthesis during DNA repair
Synthesis of DNA that proceeds from the broken 3' single-strand DNA end uses the homologous intact duplex as the template.
metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
regulation of cell cycle
Any process that modulates the rate or extent of progression through the cell cycle.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
The chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
DNA metabolic process
The chemical reactions and pathways involving DNA, deoxyribonucleic acid, one of the two main types of nucleic acid, consisting of a long, unbranched macromolecule formed from one, or more commonly, two, strands of linked deoxyribonucleotides.
DNA-dependent DNA replication
The process whereby new strands of DNA are synthesized, using parental DNA as a template for the DNA-dependent DNA polymerases that synthesize the new strands.
DNA replication initiation
The process by which DNA replication is started; this involves the separation of a stretch of the DNA double helix, the recruitment of DNA polymerases and the initiation of polymerase action.
DNA strand elongation during DNA replication
The process by which a DNA strand is synthesized from template DNA during replication by the action of polymerases, which add nucleotides to the 3' end of the nascent DNA strand.
DNA repair
The process of restoring DNA after damage. Genomes are subject to damage by chemical and physical agents in the environment (e.g. UV and ionizing radiations, chemical mutagens, fungal and bacterial toxins, etc.) and by free radicals or alkylating agents endogenously generated in metabolism. DNA is also damaged because of errors during its replication. A variety of different DNA repair pathways have been reported that include direct reversal, base excision repair, nucleotide excision repair, photoreactivation, bypass, double-strand break repair pathway, and mismatch repair pathway.
nucleotide-excision repair
In nucleotide excision repair a small region of the strand surrounding the damage is removed from the DNA helix as an oligonucleotide. The small gap left in the DNA helix is filled in by the sequential action of DNA polymerase and DNA ligase. Nucleotide excision repair recognizes a wide range of substrates, including damage caused by UV irradiation (pyrimidine dimers and 6-4 photoproducts) and chemicals (intrastrand cross-links and bulky adducts).
nucleotide-excision repair, DNA gap filling
Repair of the gap in the DNA helix by DNA polymerase and DNA ligase after the portion of the strand containing the lesion has been removed by pyrimidine-dimer repair enzymes.
response to stress
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a disturbance in organismal or cellular homeostasis, usually, but not necessarily, exogenous (e.g. temperature, humidity, ionizing radiation).
response to DNA damage stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating damage to its DNA from environmental insults or errors during metabolism.
organelle organization
A process that is carried out at the cellular level which results in the formation, arrangement of constituent parts, or disassembly of an organelle within a cell. An organelle is an organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
cell cycle
The progression of biochemical and morphological phases and events that occur in a cell during successive cell replication or nuclear replication events. Canonically, the cell cycle comprises the replication and segregation of genetic material followed by the division of the cell, but in endocycles or syncytial cells nuclear replication or nuclear division may not be followed by cell division.
mitosis
A cell cycle process comprising the steps by which the nucleus of a eukaryotic cell divides; the process involves condensation of chromosomal DNA into a highly compacted form. Canonically, mitosis produces two daughter nuclei whose chromosome complement is identical to that of the mother cell.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
biosynthetic process
The chemical reactions and pathways resulting in the formation of substances; typically the energy-requiring part of metabolism in which simpler substances are transformed into more complex ones.
macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
cellular component organization
A process that is carried out at the cellular level which results in the formation, arrangement of constituent parts, or disassembly of a cellular component.
cell cycle process
A cellular process that is involved in the progression of biochemical and morphological phases and events that occur in a cell during successive cell replication or nuclear replication events.
cell cycle phase
A cell cycle process comprising the steps by which a cell progresses through one of the biochemical and morphological phases and events that occur during successive cell replication or nuclear replication events.
DNA strand elongation
The DNA metabolic process by which a DNA strand is synthesized by adding nucleotides to the 3' end of an existing DNA stand.
cellular response to stress
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating the organism is under stress. The stress is usually, but not necessarily, exogenous (e.g. temperature, humidity, ionizing radiation).
cellular macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, carried out by individual cells.
cellular biopolymer metabolic process
The chemical reactions and pathways involving biopolymers, long, repeating chains of monomers found in nature, such as polysaccharides and proteins, as carried out by individual cells.
cellular biopolymer biosynthetic process
The chemical reactions and pathways resulting in the formation of biopolymers, long, repeating chains of monomers found in nature, such as polysaccharides and proteins, as carried out by individual cells.
cellular response to DNA damage stimulus
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating damage to its DNA from environmental insults or errors during metabolism.
macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
biopolymer metabolic process
The chemical reactions and pathways involving biopolymers, long, repeating chains of monomers found in nature, such as polysaccharides and proteins.
biopolymer biosynthetic process
The chemical reactions and pathways resulting in the formation of biopolymers, long, repeating chains of monomers found in nature e.g. polysaccharides and proteins.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
primary metabolic process
The chemical reactions and pathways involving those compounds which are formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
cellular biosynthetic process
The chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
organelle fission
The creation of two or more organelles by division of one organelle.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
response to stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
cell division
The process resulting in the physical partitioning and separation of a cell into daughter cells.
cellular response to stimulus
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
all
This term is the most general term possible
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
cellular response to stimulus
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
cellular biosynthetic process
The chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
The chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
cell cycle process
A cellular process that is involved in the progression of biochemical and morphological phases and events that occur in a cell during successive cell replication or nuclear replication events.
regulation of cell cycle
Any process that modulates the rate or extent of progression through the cell cycle.
cellular response to stress
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating the organism is under stress. The stress is usually, but not necessarily, exogenous (e.g. temperature, humidity, ionizing radiation).
cellular macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, carried out by individual cells.
biopolymer biosynthetic process
The chemical reactions and pathways resulting in the formation of biopolymers, long, repeating chains of monomers found in nature e.g. polysaccharides and proteins.
cellular macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, carried out by individual cells.
cellular biopolymer metabolic process
The chemical reactions and pathways involving biopolymers, long, repeating chains of monomers found in nature, such as polysaccharides and proteins, as carried out by individual cells.
cellular response to DNA damage stimulus
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating damage to its DNA from environmental insults or errors during metabolism.
cellular biopolymer biosynthetic process
The chemical reactions and pathways resulting in the formation of biopolymers, long, repeating chains of monomers found in nature, such as polysaccharides and proteins, as carried out by individual cells.
DNA metabolic process
The chemical reactions and pathways involving DNA, deoxyribonucleic acid, one of the two main types of nucleic acid, consisting of a long, unbranched macromolecule formed from one, or more commonly, two, strands of linked deoxyribonucleotides.
cellular biopolymer biosynthetic process
The chemical reactions and pathways resulting in the formation of biopolymers, long, repeating chains of monomers found in nature, such as polysaccharides and proteins, as carried out by individual cells.
mitosis
A cell cycle process comprising the steps by which the nucleus of a eukaryotic cell divides; the process involves condensation of chromosomal DNA into a highly compacted form. Canonically, mitosis produces two daughter nuclei whose chromosome complement is identical to that of the mother cell.
M phase of mitotic cell cycle
A cell cycle process comprising the steps by which a cell progresses through M phase, the part of the mitotic cell cycle during which mitosis takes place.
DNA repair
The process of restoring DNA after damage. Genomes are subject to damage by chemical and physical agents in the environment (e.g. UV and ionizing radiations, chemical mutagens, fungal and bacterial toxins, etc.) and by free radicals or alkylating agents endogenously generated in metabolism. DNA is also damaged because of errors during its replication. A variety of different DNA repair pathways have been reported that include direct reversal, base excision repair, nucleotide excision repair, photoreactivation, bypass, double-strand break repair pathway, and mismatch repair pathway.
DNA replication
The process whereby new strands of DNA are synthesized. The template for replication can either be an existing DNA molecule or RNA.
DNA synthesis during DNA repair
Synthesis of DNA that proceeds from the broken 3' single-strand DNA end uses the homologous intact duplex as the template.
mitosis
A cell cycle process comprising the steps by which the nucleus of a eukaryotic cell divides; the process involves condensation of chromosomal DNA into a highly compacted form. Canonically, mitosis produces two daughter nuclei whose chromosome complement is identical to that of the mother cell.
DNA replication initiation
The process by which DNA replication is started; this involves the separation of a stretch of the DNA double helix, the recruitment of DNA polymerases and the initiation of polymerase action.
DNA strand elongation during DNA replication
The process by which a DNA strand is synthesized from template DNA during replication by the action of polymerases, which add nucleotides to the 3' end of the nascent DNA strand.
nucleotide-excision repair, DNA gap filling
Repair of the gap in the DNA helix by DNA polymerase and DNA ligase after the portion of the strand containing the lesion has been removed by pyrimidine-dimer repair enzymes.

nuclear chromosome
A chromosome found in the nucleus of a eukaryotic cell.
intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
collagen
Any of the various assemblies in which collagen chains form a left-handed triple helix; may assemble into higher order structures.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
proteinaceous extracellular matrix
A layer consisting mainly of proteins (especially collagen) and glycosaminoglycans (mostly as proteoglycans) that forms a sheet underlying or overlying cells such as endothelial and epithelial cells. The proteins are secreted by cells in the vicinity.
collagen type IV
A collagen heterotrimer containing type IV alpha chains; [alpha1(IV)]2alpha2(IV) trimers are commonly observed, although more type IV alpha chains exist and may be present in type IV trimers; type IV collagen triple helices associate to form nets within basement membranes.
basement membrane
A thin layer of dense material found in various animal tissues interposed between the cells and the adjacent connective tissue. It consists of the basal lamina plus an associated layer of reticulin fibers.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
nucleus
A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent.
nucleoplasm
That part of the nuclear content other than the chromosomes or the nucleolus.
replication fork
The Y-shaped region of a replicating DNA molecule, resulting from the separation of the DNA strands and in which the synthesis of new strands takes place. Also includes associated protein complexes.
alpha DNA polymerase:primase complex
A complex of four polypeptides, comprising large and small DNA polymerase alpha subunits and two primase subunits, which catalyzes the synthesis of an RNA primer on the lagging strand of replicating DNA; the smaller of the two primase subunits alone can catalyze oligoribonucleotide synthesis.
chromosome
A structure composed of a very long molecule of DNA and associated proteins (e.g. histones) that carries hereditary information.
replisome
A multi-component enzymatic machine at the replication fork which mediates DNA replication. Includes DNA primase, one or more DNA polymerases, DNA helicases, and other proteins.
sheet-forming collagen
Any collagen polymer in which collagen triple helices associate to form sheet-like networks.
extracellular matrix
A structure lying external to one or more cells, which provides structural support for cells or tissues; may be completely external to the cell (as in animals) or be part of the cell (as in plants).
membrane-enclosed lumen
The enclosed volume within a sealed membrane or between two sealed membranes. Encompasses the volume enclosed by the membranes of a particular organelle, e.g. endoplasmic reticulum lumen, or the space between the two lipid bilayers of a double membrane surrounding an organelle, e.g. nuclear envelope lumen.
nuclear lumen
The volume enclosed by the nuclear inner membrane.
macromolecular complex
A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which the constituent parts function together.
protein-DNA complex
A macromolecular complex containing both protein and DNA molecules.
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane. Includes ribosomes, the cytoskeleton and chromosomes.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane and occurring within the cell. Includes ribosomes, the cytoskeleton and chromosomes.
organelle lumen
The internal volume enclosed by the membranes of a particular organelle; includes the volume enclosed by a single organelle membrane, e.g. endoplasmic reticulum lumen, or the volume enclosed by the innermost of the two lipid bilayers of an organelle envelope, e.g. nuclear lumen.
protein complex
Any macromolecular complex composed of two or more polypeptide subunits, which may or may not be identical. Protein complexes may have other associated non-protein prosthetic groups, such as nucleotides, metal ions or carbohydrate groups.
nuclear replication fork
The Y-shaped region of a nuclear replicating DNA molecule, resulting from the separation of the DNA strands and in which the synthesis of new strands takes place. Also includes associated protein complexes.
nuclear replisome
A multi-component enzymatic machine at the nuclear replication fork, which mediates DNA replication. Includes DNA primase, one or more DNA polymerases, DNA helicases, and other proteins.
extracellular matrix part
Any constituent part of the extracellular matrix, the structure lying external to one or more cells, which provides structural support for cells or tissues; may be completely external to the cell (as in animals) or be part of the cell (as often seen in plants).
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
chromosomal part
Any constituent part of a chromosome, a structure composed of a very long molecule of DNA and associated proteins (e.g. histones) that carries hereditary information.
nuclear part
Any constituent part of the nucleus, a membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
nuclear chromosome part
Any constituent part of a nuclear chromosome, a chromosome found in the nucleus of a eukaryotic cell.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
intracellular organelle lumen
An organelle lumen that is part of an intracellular organelle.
all
This term is the most general term possible
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
organelle lumen
The internal volume enclosed by the membranes of a particular organelle; includes the volume enclosed by a single organelle membrane, e.g. endoplasmic reticulum lumen, or the volume enclosed by the innermost of the two lipid bilayers of an organelle envelope, e.g. nuclear lumen.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane and occurring within the cell. Includes ribosomes, the cytoskeleton and chromosomes.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
extracellular matrix part
Any constituent part of the extracellular matrix, the structure lying external to one or more cells, which provides structural support for cells or tissues; may be completely external to the cell (as in animals) or be part of the cell (as often seen in plants).
intracellular organelle lumen
An organelle lumen that is part of an intracellular organelle.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
collagen
Any of the various assemblies in which collagen chains form a left-handed triple helix; may assemble into higher order structures.
basement membrane
A thin layer of dense material found in various animal tissues interposed between the cells and the adjacent connective tissue. It consists of the basal lamina plus an associated layer of reticulin fibers.
replisome
A multi-component enzymatic machine at the replication fork which mediates DNA replication. Includes DNA primase, one or more DNA polymerases, DNA helicases, and other proteins.
nuclear lumen
The volume enclosed by the nuclear inner membrane.
nuclear chromosome part
Any constituent part of a nuclear chromosome, a chromosome found in the nucleus of a eukaryotic cell.
nucleoplasm
That part of the nuclear content other than the chromosomes or the nucleolus.
alpha DNA polymerase:primase complex
A complex of four polypeptides, comprising large and small DNA polymerase alpha subunits and two primase subunits, which catalyzes the synthesis of an RNA primer on the lagging strand of replicating DNA; the smaller of the two primase subunits alone can catalyze oligoribonucleotide synthesis.
nuclear part
Any constituent part of the nucleus, a membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated.
nuclear chromosome
A chromosome found in the nucleus of a eukaryotic cell.
chromosomal part
Any constituent part of a chromosome, a structure composed of a very long molecule of DNA and associated proteins (e.g. histones) that carries hereditary information.
collagen type IV
A collagen heterotrimer containing type IV alpha chains; [alpha1(IV)]2alpha2(IV) trimers are commonly observed, although more type IV alpha chains exist and may be present in type IV trimers; type IV collagen triple helices associate to form nets within basement membranes.
replisome
A multi-component enzymatic machine at the replication fork which mediates DNA replication. Includes DNA primase, one or more DNA polymerases, DNA helicases, and other proteins.
alpha DNA polymerase:primase complex
A complex of four polypeptides, comprising large and small DNA polymerase alpha subunits and two primase subunits, which catalyzes the synthesis of an RNA primer on the lagging strand of replicating DNA; the smaller of the two primase subunits alone can catalyze oligoribonucleotide synthesis.
nuclear replication fork
The Y-shaped region of a nuclear replicating DNA molecule, resulting from the separation of the DNA strands and in which the synthesis of new strands takes place. Also includes associated protein complexes.
nuclear replisome
A multi-component enzymatic machine at the nuclear replication fork, which mediates DNA replication. Includes DNA primase, one or more DNA polymerases, DNA helicases, and other proteins.
nuclear chromosome part
Any constituent part of a nuclear chromosome, a chromosome found in the nucleus of a eukaryotic cell.
nuclear replisome
A multi-component enzymatic machine at the nuclear replication fork, which mediates DNA replication. Includes DNA primase, one or more DNA polymerases, DNA helicases, and other proteins.

| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 03030 | 2.948e-13 | 1.068 | 16 | 33 | DNA replication |
| 04110 | 1.484e-03 | 3.172 | 13 | 98 | Cell cycle |
| 03430 | 5.066e-03 | 0.7121 | 6 | 22 | Mismatch repair |
| 03410 | 2.199e-02 | 0.971 | 6 | 30 | Base excision repair |
ABHD4abhydrolase domain containing 4 (218581_at), score: 0.7 ANK2ankyrin 2, neuronal (202920_at), score: 0.68 ANKRD1ankyrin repeat domain 1 (cardiac muscle) (206029_at), score: 0.73 ANXA3annexin A3 (209369_at), score: 0.67 ARL4CADP-ribosylation factor-like 4C (202207_at), score: 0.7 ASF1BASF1 anti-silencing function 1 homolog B (S. cerevisiae) (218115_at), score: -0.81 ASPMasp (abnormal spindle) homolog, microcephaly associated (Drosophila) (219918_s_at), score: -0.75 ATF3activating transcription factor 3 (202672_s_at), score: 0.72 ATXN1ataxin 1 (203232_s_at), score: 0.69 AURKBaurora kinase B (209464_at), score: -0.8 B3GALT2UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 (217452_s_at), score: 0.65 BARD1BRCA1 associated RING domain 1 (205345_at), score: -0.83 BIRC5baculoviral IAP repeat-containing 5 (202095_s_at), score: -0.74 BRCA1breast cancer 1, early onset (204531_s_at), score: -0.79 BST1bone marrow stromal cell antigen 1 (205715_at), score: 0.87 C17orf91chromosome 17 open reading frame 91 (214696_at), score: 0.66 C1orf54chromosome 1 open reading frame 54 (219506_at), score: 0.8 C21orf45chromosome 21 open reading frame 45 (219004_s_at), score: -0.86 C4orf18chromosome 4 open reading frame 18 (219872_at), score: 0.7 C7orf10chromosome 7 open reading frame 10 (219655_at), score: 0.63 C9orf40chromosome 9 open reading frame 40 (218904_s_at), score: -0.76 CCL2chemokine (C-C motif) ligand 2 (216598_s_at), score: 0.92 CCNA2cyclin A2 (203418_at), score: -0.76 CCNE2cyclin E2 (205034_at), score: -0.83 CDC25Acell division cycle 25 homolog A (S. pombe) (204695_at), score: -0.76 CDC45LCDC45 cell division cycle 45-like (S. cerevisiae) (204126_s_at), score: -0.77 CDCA8cell division cycle associated 8 (221520_s_at), score: -0.78 CDH13cadherin 13, H-cadherin (heart) (204726_at), score: 0.79 CDK2cyclin-dependent kinase 2 (204252_at), score: -0.83 CDT1chromatin licensing and DNA replication factor 1 (209832_s_at), score: -0.89 CENPMcentromere protein M (218741_at), score: -0.78 CFHR1complement factor H-related 1 (215388_s_at), score: 0.63 CHAF1Achromatin assembly factor 1, subunit A (p150) (214426_x_at), score: -0.83 CHAF1Bchromatin assembly factor 1, subunit B (p60) (204775_at), score: -0.76 CITcitron (rho-interacting, serine/threonine kinase 21) (212801_at), score: -0.78 CKAP2cytoskeleton associated protein 2 (218252_at), score: -0.79 CLEC3BC-type lectin domain family 3, member B (205200_at), score: 0.74 CLIP3CAP-GLY domain containing linker protein 3 (212358_at), score: 0.62 COBLL1COBL-like 1 (203642_s_at), score: 0.78 COL4A1collagen, type IV, alpha 1 (211981_at), score: 0.76 COL4A2collagen, type IV, alpha 2 (211966_at), score: 0.64 COL4A5collagen, type IV, alpha 5 (213110_s_at), score: 0.88 CORINcorin, serine peptidase (220356_at), score: 0.82 CPA3carboxypeptidase A3 (mast cell) (205624_at), score: 0.7 CYFIP2cytoplasmic FMR1 interacting protein 2 (215785_s_at), score: 0.88 CYP1B1cytochrome P450, family 1, subfamily B, polypeptide 1 (202436_s_at), score: 0.66 DAAM2dishevelled associated activator of morphogenesis 2 (212793_at), score: 0.85 DACT1dapper, antagonist of beta-catenin, homolog 1 (Xenopus laevis) (219179_at), score: 0.73 DBNDD2dysbindin (dystrobrevin binding protein 1) domain containing 2 (218094_s_at), score: 0.7 DCKdeoxycytidine kinase (203302_at), score: -0.76 DDX11DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 11 (CHL1-like helicase homolog, S. cerevisiae) (208149_x_at), score: -0.76 DHCR77-dehydrocholesterol reductase (201790_s_at), score: 0.63 DLEU2Ldeleted in lymphocytic leukemia 2-like (215629_s_at), score: -0.75 DMDdystrophin (203881_s_at), score: 0.94 DRAMdamage-regulated autophagy modulator (218627_at), score: 0.64 DSN1DSN1, MIND kinetochore complex component, homolog (S. cerevisiae) (219512_at), score: -0.79 DTLdenticleless homolog (Drosophila) (218585_s_at), score: -0.82 DVL3dishevelled, dsh homolog 3 (Drosophila) (201908_at), score: 0.62 EDIL3EGF-like repeats and discoidin I-like domains 3 (207379_at), score: 0.8 EFHD1EF-hand domain family, member D1 (209343_at), score: 0.63 EMP2epithelial membrane protein 2 (204975_at), score: -0.77 EMX2empty spiracles homeobox 2 (221950_at), score: -0.81 ENPP1ectonucleotide pyrophosphatase/phosphodiesterase 1 (205066_s_at), score: -0.93 ERCC6Lexcision repair cross-complementing rodent repair deficiency, complementation group 6-like (219650_at), score: -0.75 EXO1exonuclease 1 (204603_at), score: -0.76 EXOSC9exosome component 9 (205061_s_at), score: -0.83 EZH2enhancer of zeste homolog 2 (Drosophila) (203358_s_at), score: -0.85 FAM8A1family with sequence similarity 8, member A1 (203420_at), score: 0.69 FANCGFanconi anemia, complementation group G (203564_at), score: -0.79 FEN1flap structure-specific endonuclease 1 (204768_s_at), score: -0.78 FOXM1forkhead box M1 (202580_x_at), score: -0.75 FOXN3forkhead box N3 (218031_s_at), score: 0.64 FRMD4AFERM domain containing 4A (208476_s_at), score: 0.71 FRYfurry homolog (Drosophila) (204072_s_at), score: 0.73 FYCO1FYVE and coiled-coil domain containing 1 (218204_s_at), score: 0.76 GABBR2gamma-aminobutyric acid (GABA) B receptor, 2 (209990_s_at), score: -0.77 GALNT3UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3) (203397_s_at), score: 0.78 GDF15growth differentiation factor 15 (221577_x_at), score: 0.69 GINS1GINS complex subunit 1 (Psf1 homolog) (206102_at), score: -0.84 GINS2GINS complex subunit 2 (Psf2 homolog) (221521_s_at), score: -0.79 GINS4GINS complex subunit 4 (Sld5 homolog) (211767_at), score: -0.77 GNPTABN-acetylglucosamine-1-phosphate transferase, alpha and beta subunits (212959_s_at), score: 0.62 GPERG protein-coupled estrogen receptor 1 (210640_s_at), score: -0.9 GPR116G protein-coupled receptor 116 (212950_at), score: 0.65 GRAMD1CGRAM domain containing 1C (219313_at), score: -0.78 GSTM1glutathione S-transferase mu 1 (204550_x_at), score: 0.62 GSTM3glutathione S-transferase mu 3 (brain) (202554_s_at), score: 0.7 GTDC1glycosyltransferase-like domain containing 1 (219770_at), score: 0.69 GTSE1G-2 and S-phase expressed 1 (204318_s_at), score: -0.77 HIVEP1human immunodeficiency virus type I enhancer binding protein 1 (204512_at), score: 0.81 HNMThistamine N-methyltransferase (204112_s_at), score: 0.8 HOXA9homeobox A9 (214651_s_at), score: -0.79 HS3ST3A1heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1 (219985_at), score: -0.81 ICAM1intercellular adhesion molecule 1 (202638_s_at), score: 0.69 IL6interleukin 6 (interferon, beta 2) (205207_at), score: 0.7 ITGA8integrin, alpha 8 (214265_at), score: 0.66 JAG1jagged 1 (Alagille syndrome) (216268_s_at), score: 0.97 KIF15kinesin family member 15 (219306_at), score: -0.78 KIF18Bkinesin family member 18B (222039_at), score: -0.78 KIF4Akinesin family member 4A (218355_at), score: -0.75 KLF9Kruppel-like factor 9 (203543_s_at), score: 0.75 KNTC1kinetochore associated 1 (206316_s_at), score: -0.78 KRT18keratin 18 (201596_x_at), score: 0.66 LAMA5laminin, alpha 5 (210150_s_at), score: 0.75 LGALS3BPlectin, galactoside-binding, soluble, 3 binding protein (200923_at), score: 0.93 LIFleukemia inhibitory factor (cholinergic differentiation factor) (205266_at), score: 0.64 LIG1ligase I, DNA, ATP-dependent (202726_at), score: -0.84 LIPGlipase, endothelial (219181_at), score: 0.68 LMBRD1LMBR1 domain containing 1 (218191_s_at), score: 0.64 LMCD1LIM and cysteine-rich domains 1 (218574_s_at), score: 0.73 LOC731884similar to programmed cell death 6 interacting protein (217520_x_at), score: -0.78 LPHN2latrophilin 2 (206953_s_at), score: 0.78 LPPR4plasticity related gene 1 (213496_at), score: 0.82 LPXNleupaxin (216250_s_at), score: -0.8 LRP5low density lipoprotein receptor-related protein 5 (209468_at), score: 0.69 LRRC15leucine rich repeat containing 15 (213909_at), score: -0.85 LRRC17leucine rich repeat containing 17 (205381_at), score: 0.93 LRRC32leucine rich repeat containing 32 (203835_at), score: 0.75 MANSC1MANSC domain containing 1 (220945_x_at), score: 0.69 MAP3K8mitogen-activated protein kinase kinase kinase 8 (205027_s_at), score: 0.75 MARCH2membrane-associated ring finger (C3HC4) 2 (210075_at), score: 0.68 MBPmyelin basic protein (210136_at), score: 0.93 MCM10minichromosome maintenance complex component 10 (220651_s_at), score: -0.86 MCM2minichromosome maintenance complex component 2 (202107_s_at), score: -0.81 MCM3minichromosome maintenance complex component 3 (201555_at), score: -0.8 MCM4minichromosome maintenance complex component 4 (212141_at), score: -0.89 MCM5minichromosome maintenance complex component 5 (216237_s_at), score: -0.84 MCM7minichromosome maintenance complex component 7 (210983_s_at), score: -0.79 MESTmesoderm specific transcript homolog (mouse) (202016_at), score: 0.69 MGPmatrix Gla protein (202291_s_at), score: 0.77 MKI67antigen identified by monoclonal antibody Ki-67 (212022_s_at), score: -0.76 MMP3matrix metallopeptidase 3 (stromelysin 1, progelatinase) (205828_at), score: -0.83 MYBL1v-myb myeloblastosis viral oncogene homolog (avian)-like 1 (213906_at), score: -0.96 MYO1Emyosin IE (203072_at), score: 0.75 NCAPG2non-SMC condensin II complex, subunit G2 (219588_s_at), score: -0.78 NCAPHnon-SMC condensin I complex, subunit H (212949_at), score: -0.76 NDUFA4L2NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2 (218484_at), score: 0.73 NEDD9neural precursor cell expressed, developmentally down-regulated 9 (202149_at), score: 0.87 NID2nidogen 2 (osteonidogen) (204114_at), score: 0.68 NRG1neuregulin 1 (206343_s_at), score: -0.86 NRIP3nuclear receptor interacting protein 3 (219557_s_at), score: -0.8 NRXN3neurexin 3 (205795_at), score: 0.91 NUAK1NUAK family, SNF1-like kinase, 1 (204589_at), score: 0.67 OLFM1olfactomedin 1 (213131_at), score: -0.82 OPN3opsin 3 (219032_x_at), score: 0.62 OPTNoptineurin (202073_at), score: 0.75 ORC6Lorigin recognition complex, subunit 6 like (yeast) (219105_x_at), score: -0.86 OSBPL10oxysterol binding protein-like 10 (219073_s_at), score: 0.63 OSMRoncostatin M receptor (205729_at), score: 0.67 PDCD4programmed cell death 4 (neoplastic transformation inhibitor) (202731_at), score: 0.71 PDGFAplatelet-derived growth factor alpha polypeptide (205463_s_at), score: 0.85 PDGFRBplatelet-derived growth factor receptor, beta polypeptide (202273_at), score: 0.81 PDGFRLplatelet-derived growth factor receptor-like (205226_at), score: 0.75 PDLIM3PDZ and LIM domain 3 (209621_s_at), score: 0.65 PENKproenkephalin (213791_at), score: -0.75 PIRpirin (iron-binding nuclear protein) (207469_s_at), score: 0.67 PKMYT1protein kinase, membrane associated tyrosine/threonine 1 (204267_x_at), score: -0.85 PLA2G16phospholipase A2, group XVI (209581_at), score: 0.76 PLK4polo-like kinase 4 (Drosophila) (204887_s_at), score: -0.75 PLSCR4phospholipid scramblase 4 (218901_at), score: 0.79 PLXND1plexin D1 (38671_at), score: 0.73 PNMA2paraneoplastic antigen MA2 (209598_at), score: 0.81 POLA1polymerase (DNA directed), alpha 1, catalytic subunit (204835_at), score: -0.86 POLA2polymerase (DNA directed), alpha 2 (70kD subunit) (204441_s_at), score: -0.76 POLD1polymerase (DNA directed), delta 1, catalytic subunit 125kDa (203422_at), score: -0.85 POLD3polymerase (DNA-directed), delta 3, accessory subunit (212836_at), score: -0.82 POLE2polymerase (DNA directed), epsilon 2 (p59 subunit) (205909_at), score: -0.78 POSTNperiostin, osteoblast specific factor (210809_s_at), score: 0.67 PRIM1primase, DNA, polypeptide 1 (49kDa) (205053_at), score: -0.8 PRKAG2protein kinase, AMP-activated, gamma 2 non-catalytic subunit (218292_s_at), score: 0.74 PRKG1protein kinase, cGMP-dependent, type I (207119_at), score: 0.65 PROS1protein S (alpha) (207808_s_at), score: 0.69 PRRX2paired related homeobox 2 (219729_at), score: -0.75 PSMB9proteasome (prosome, macropain) subunit, beta type, 9 (large multifunctional peptidase 2) (204279_at), score: 0.63 PSMC3IPPSMC3 interacting protein (213951_s_at), score: -0.82 PTGER4prostaglandin E receptor 4 (subtype EP4) (204897_at), score: 0.74 PTPRZ1protein tyrosine phosphatase, receptor-type, Z polypeptide 1 (204469_at), score: 0.7 RAB33ARAB33A, member RAS oncogene family (206039_at), score: 0.63 RAD54BRAD54 homolog B (S. cerevisiae) (219494_at), score: -0.91 RAD54LRAD54-like (S. cerevisiae) (204558_at), score: -0.76 RBL1retinoblastoma-like 1 (p107) (205296_at), score: -0.78 RELNreelin (205923_at), score: 0.84 RFC3replication factor C (activator 1) 3, 38kDa (204128_s_at), score: -0.83 RFC4replication factor C (activator 1) 4, 37kDa (204023_at), score: -0.77 RFWD3ring finger and WD repeat domain 3 (218564_at), score: -0.83 RMI1RMI1, RecQ mediated genome instability 1, homolog (S. cerevisiae) (218979_at), score: -0.76 RNASEH2Aribonuclease H2, subunit A (203022_at), score: -0.81 RNASET2ribonuclease T2 (217983_s_at), score: 0.72 RTN1reticulon 1 (203485_at), score: 0.72 RWDD2ARWD domain containing 2A (213555_at), score: 0.64 S100A2S100 calcium binding protein A2 (204268_at), score: -0.81 SCARA3scavenger receptor class A, member 3 (219416_at), score: -0.92 SCN9Asodium channel, voltage-gated, type IX, alpha subunit (206950_at), score: 0.8 SECTM1secreted and transmembrane 1 (213716_s_at), score: 0.64 SERPINB2serpin peptidase inhibitor, clade B (ovalbumin), member 2 (204614_at), score: 0.65 SLC25A4solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 4 (202825_at), score: 0.63 SLC46A3solute carrier family 46, member 3 (214719_at), score: 0.78 SMC2structural maintenance of chromosomes 2 (204240_s_at), score: -0.78 SNTB1syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1) (208608_s_at), score: 0.74 SOBPsine oculis binding protein homolog (Drosophila) (218974_at), score: 0.72 SPOCK1sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1 (202363_at), score: 0.67 SPSB1splA/ryanodine receptor domain and SOCS box containing 1 (219677_at), score: 0.69 SQLEsqualene epoxidase (213562_s_at), score: 0.65 SQRDLsulfide quinone reductase-like (yeast) (217995_at), score: 0.7 SSX2IPsynovial sarcoma, X breakpoint 2 interacting protein (203016_s_at), score: -0.93 STAT1signal transducer and activator of transcription 1, 91kDa (AFFX-HUMISGF3A/M97935_MA_at), score: 0.7 STAT4signal transducer and activator of transcription 4 (206118_at), score: 0.87 STEAP1six transmembrane epithelial antigen of the prostate 1 (205542_at), score: -0.8 STILSCL/TAL1 interrupting locus (205339_at), score: -0.78 STK38Lserine/threonine kinase 38 like (212572_at), score: 0.88 SUV39H1suppressor of variegation 3-9 homolog 1 (Drosophila) (218619_s_at), score: -0.76 SYNE1spectrin repeat containing, nuclear envelope 1 (209447_at), score: 0.62 SYNGR2synaptogyrin 2 (201079_at), score: 0.77 TACC3transforming, acidic coiled-coil containing protein 3 (218308_at), score: -0.79 TCF7L1transcription factor 7-like 1 (T-cell specific, HMG-box) (221016_s_at), score: 0.8 TELO2TEL2, telomere maintenance 2, homolog (S. cerevisiae) (34260_at), score: -0.75 TFPItissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) (209676_at), score: 0.83 THBS2thrombospondin 2 (203083_at), score: 0.62 TINAGL1tubulointerstitial nephritis antigen-like 1 (219058_x_at), score: 0.75 TMEM194Atransmembrane protein 194A (212621_at), score: -0.87 TNFAIP8tumor necrosis factor, alpha-induced protein 8 (210260_s_at), score: 0.62 TNFRSF10Btumor necrosis factor receptor superfamily, member 10b (209295_at), score: 0.68 TNFSF4tumor necrosis factor (ligand) superfamily, member 4 (207426_s_at), score: 0.77 TNXAtenascin XA pseudogene (213451_x_at), score: 0.99 TNXBtenascin XB (216333_x_at), score: 1 TPCN1two pore segment channel 1 (217914_at), score: 0.63 TPD52L1tumor protein D52-like 1 (203786_s_at), score: 0.67 TRIM22tripartite motif-containing 22 (213293_s_at), score: 0.71 TRIP13thyroid hormone receptor interactor 13 (204033_at), score: -0.77 TTKTTK protein kinase (204822_at), score: -0.77 TUFT1tuftelin 1 (205807_s_at), score: 0.72 UBA7ubiquitin-like modifier activating enzyme 7 (203281_s_at), score: 0.76 UNGuracil-DNA glycosylase (202330_s_at), score: -0.79 VRK1vaccinia related kinase 1 (203856_at), score: -0.76 WDR62WD repeat domain 62 (215218_s_at), score: -0.75 WDR76WD repeat domain 76 (205519_at), score: -0.81 WHSC1Wolf-Hirschhorn syndrome candidate 1 (209053_s_at), score: -0.85 WRAP53WD repeat containing, antisense to TP53 (44563_at), score: -0.79 ZNF423zinc finger protein 423 (214761_at), score: 0.82 ZNF536zinc finger protein 536 (206403_at), score: -0.85 ZNF85zinc finger protein 85 (206572_x_at), score: -0.75
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-3860-raw-cel-1561690344.cel | 10 | 5 | HGPS | hgu133a | none | GM00038C |
| E-GEOD-3860-raw-cel-1561690472.cel | 17 | 5 | HGPS | hgu133a | none | GM00038C |
| E-GEOD-3860-raw-cel-1561690223.cel | 3 | 5 | HGPS | hgu133a | none | GM00038C |
| E-GEOD-3860-raw-cel-1561690392.cel | 14 | 5 | HGPS | hgu133a | none | GMO8398C |
| E-GEOD-3860-raw-cel-1561690256.cel | 6 | 5 | HGPS | hgu133a | none | GMO8398C |
| E-GEOD-3860-raw-cel-1561690304.cel | 8 | 5 | HGPS | hgu133a | none | GMO8398C |
| 5CTwin.CEL | 5 | 2 | DS-twin | hgu133plus2 | Down | DS-twin 5 |
| E-GEOD-3860-raw-cel-1561690360.cel | 12 | 5 | HGPS | hgu133a | none | GM0316B |
| 6Twin.CEL | 6 | 2 | DS-twin | hgu133plus2 | none | DS-twin 6 |
| E-GEOD-3860-raw-cel-1561690416.cel | 15 | 5 | HGPS | hgu133a | none | GM0316B |
| E-GEOD-3407-raw-cel-1437949557.cel | 1 | 4 | Cockayne | hgu133a | CS | eGFP |
| 1Twin.CEL | 1 | 2 | DS-twin | hgu133plus2 | Down | DS-twin 1 |
| 2Twin.CEL | 2 | 2 | DS-twin | hgu133plus2 | none | DS-twin 2 |
| E-GEOD-4219-raw-cel-1311956418.cel | 13 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| 47B.CEL | 4 | 3 | DS-mosaic | hgu133plus2 | Down mosaic | DS-mosaic 4 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690432.cel | 16 | 5 | HGPS | hgu133a | HGPS | AG10750 |
| E-GEOD-3860-raw-cel-1561690231.cel | 4 | 5 | HGPS | hgu133a | HGPS | AG10750 |
| 46A.CEL | 1 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 1 |
| E-TABM-263-raw-cel-1515486351.cel | 36 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486291.cel | 33 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690336.cel | 9 | 5 | HGPS | hgu133a | HGPS | AG10750 |
| E-TABM-263-raw-cel-1515486271.cel | 32 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485851.cel | 11 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486331.cel | 35 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486231.cel | 30 | 6 | Cycle | hgu133a2 | none | Cycle 1 |