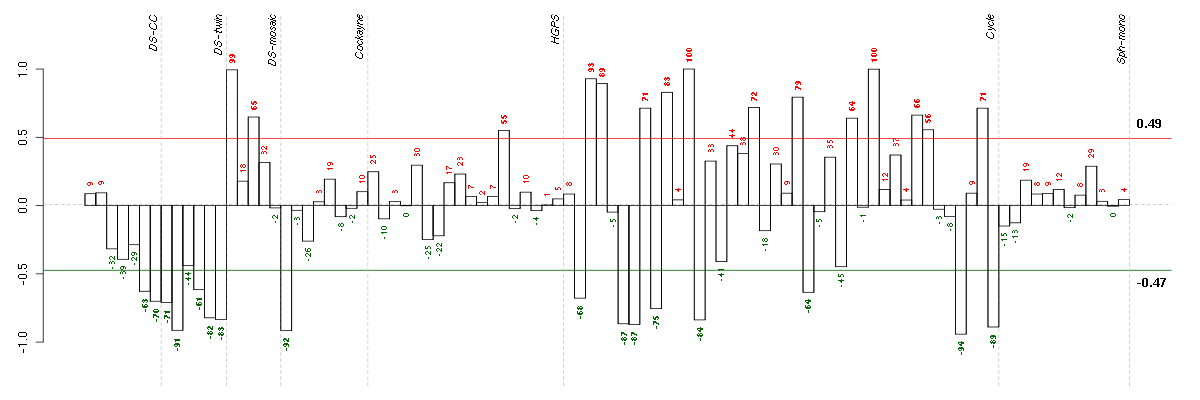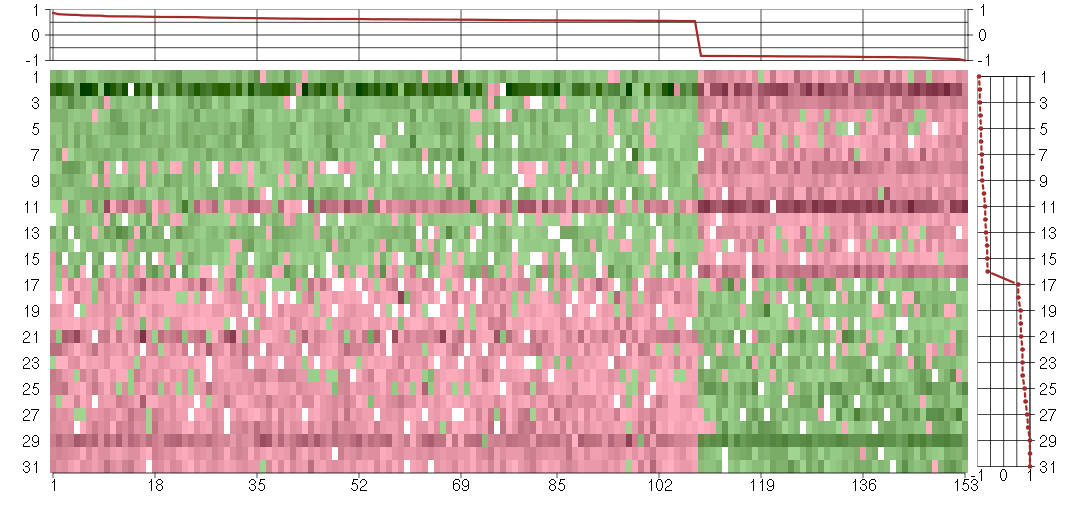

Under-expression is coded with green,
over-expression with red color.

cell adhesion
The attachment of a cell, either to another cell or to an underlying substrate such as the extracellular matrix, via cell adhesion molecules.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
biological adhesion
The attachment of a cell or organism to a substrate or other organism.
all
This term is the most general term possible
cell adhesion
The attachment of a cell, either to another cell or to an underlying substrate such as the extracellular matrix, via cell adhesion molecules.


ABCD1ATP-binding cassette, sub-family D (ALD), member 1 (205142_x_at), score: 0.66 ABLIM3actin binding LIM protein family, member 3 (205730_s_at), score: 0.61 ACTN3actinin, alpha 3 (206891_at), score: -0.83 ALDH1L1aldehyde dehydrogenase 1 family, member L1 (215798_at), score: -0.93 ANXA8L2annexin A8-like 2 (203074_at), score: -0.84 ARMC9armadillo repeat containing 9 (219637_at), score: 0.56 ARSAarylsulfatase A (204443_at), score: 0.73 ATF5activating transcription factor 5 (204999_s_at), score: 0.56 ATP6V0A1ATPase, H+ transporting, lysosomal V0 subunit a1 (205095_s_at), score: 0.57 BAT2HLA-B associated transcript 2 (212081_x_at), score: 0.68 BAT2LHLA-B associated transcript 2-like (212068_s_at), score: 0.57 BCL2L1BCL2-like 1 (215037_s_at), score: 0.57 BDH23-hydroxybutyrate dehydrogenase, type 2 (218285_s_at), score: 0.55 C16orf45chromosome 16 open reading frame 45 (212736_at), score: 0.61 CALB2calbindin 2 (205428_s_at), score: -0.83 CALCOCO1calcium binding and coiled-coil domain 1 (209002_s_at), score: 0.67 CCDC41coiled-coil domain containing 41 (219644_at), score: -0.82 CDAcytidine deaminase (205627_at), score: 0.57 CHRNA10cholinergic receptor, nicotinic, alpha 10 (220210_at), score: -0.85 CLIP3CAP-GLY domain containing linker protein 3 (212358_at), score: 0.67 COL11A2collagen, type XI, alpha 2 (216993_s_at), score: -0.9 COL18A1collagen, type XVIII, alpha 1 (209081_s_at), score: 0.71 CORO1Bcoronin, actin binding protein, 1B (64486_at), score: 0.55 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: 0.6 CTSEcathepsin E (205927_s_at), score: -0.85 DDR1discoidin domain receptor tyrosine kinase 1 (208779_x_at), score: 0.57 DEAF1deformed epidermal autoregulatory factor 1 (Drosophila) (209407_s_at), score: 0.56 DENND3DENN/MADD domain containing 3 (212974_at), score: 0.59 DGCR2DiGeorge syndrome critical region gene 2 (214198_s_at), score: -0.86 DMPKdystrophia myotonica-protein kinase (37996_s_at), score: 0.73 DNAJC4DnaJ (Hsp40) homolog, subfamily C, member 4 (206782_s_at), score: 0.61 DOPEY1dopey family member 1 (40612_at), score: -0.89 DRG2developmentally regulated GTP binding protein 2 (203267_s_at), score: 0.56 DSPPdentin sialophosphoprotein (221681_s_at), score: -0.85 DUS2Ldihydrouridine synthase 2-like, SMM1 homolog (S. cerevisiae) (47105_at), score: -0.84 ECE1endothelin converting enzyme 1 (201750_s_at), score: 0.6 ECM1extracellular matrix protein 1 (209365_s_at), score: 0.76 ENTPD1ectonucleoside triphosphate diphosphohydrolase 1 (209473_at), score: -0.84 EPHA4EPH receptor A4 (206114_at), score: -0.82 EPHX1epoxide hydrolase 1, microsomal (xenobiotic) (202017_at), score: 0.55 FAM102Afamily with sequence similarity 102, member A (212400_at), score: 0.58 FBXL14F-box and leucine-rich repeat protein 14 (213145_at), score: 0.6 FBXO2F-box protein 2 (219305_x_at), score: -0.83 FCER1GFc fragment of IgE, high affinity I, receptor for; gamma polypeptide (204232_at), score: -0.86 FETUBfetuin B (214417_s_at), score: -0.88 FHOD1formin homology 2 domain containing 1 (218530_at), score: 0.76 FLJ40113golgi autoantigen, golgin subfamily a-like pseudogene (213212_x_at), score: -0.82 FOXC2forkhead box C2 (MFH-1, mesenchyme forkhead 1) (214520_at), score: 0.64 FOXF1forkhead box F1 (205935_at), score: 0.6 FOXK2forkhead box K2 (203064_s_at), score: 0.66 GM2AGM2 ganglioside activator (212737_at), score: 0.55 GMPRguanosine monophosphate reductase (204187_at), score: 0.63 GOLT1Bgolgi transport 1 homolog B (S. cerevisiae) (218193_s_at), score: -0.82 HAB1B1 for mucin (215778_x_at), score: -0.92 HAMPhepcidin antimicrobial peptide (220491_at), score: -0.91 HBS1LHBS1-like (S. cerevisiae) (209314_s_at), score: -0.85 HHEXhematopoietically expressed homeobox (215933_s_at), score: 0.63 HLA-Emajor histocompatibility complex, class I, E (200904_at), score: 0.79 HSF1heat shock transcription factor 1 (202344_at), score: 0.63 IDUAiduronidase, alpha-L- (205059_s_at), score: 0.8 IGHMBP2immunoglobulin mu binding protein 2 (215980_s_at), score: -0.84 IL9Rinterleukin 9 receptor (217212_s_at), score: -0.94 INE1inactivation escape 1 (non-protein coding) (207252_at), score: -0.86 INPP1inositol polyphosphate-1-phosphatase (202794_at), score: 0.64 JUNDjun D proto-oncogene (203751_x_at), score: 0.71 JUPjunction plakoglobin (201015_s_at), score: 0.73 KAL1Kallmann syndrome 1 sequence (205206_at), score: -0.87 KIAA1305KIAA1305 (220911_s_at), score: 0.72 KLHL26kelch-like 26 (Drosophila) (219354_at), score: 0.66 LEPREL2leprecan-like 2 (204854_at), score: 0.64 LITAFlipopolysaccharide-induced TNF factor (200706_s_at), score: 0.55 MAN1C1mannosidase, alpha, class 1C, member 1 (218918_at), score: 0.79 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: 0.81 MAP2K3mitogen-activated protein kinase kinase 3 (207667_s_at), score: 0.7 MAP3K6mitogen-activated protein kinase kinase kinase 6 (219278_at), score: 0.58 MEOX1mesenchyme homeobox 1 (205619_s_at), score: -0.86 MYL4myosin, light chain 4, alkali; atrial, embryonic (210088_x_at), score: -0.87 NACAP1nascent-polypeptide-associated complex alpha polypeptide pseudogene 1 (211445_x_at), score: -0.88 NCKAP1NCK-associated protein 1 (207738_s_at), score: -0.83 NCRNA00093non-protein coding RNA 93 (210723_x_at), score: -0.84 NFKB1nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 (209239_at), score: 0.71 NFKBIAnuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha (201502_s_at), score: 0.59 NID1nidogen 1 (202008_s_at), score: 0.56 NINJ1ninjurin 1 (203045_at), score: 0.77 NOVA2neuro-oncological ventral antigen 2 (206477_s_at), score: -0.87 OR7E47Polfactory receptor, family 7, subfamily E, member 47 pseudogene (216698_x_at), score: -0.86 PARVBparvin, beta (37965_at), score: 0.57 PCDHG@protocadherin gamma cluster (215836_s_at), score: 0.68 PCDHGA11protocadherin gamma subfamily A, 11 (211876_x_at), score: 0.71 PCDHGA3protocadherin gamma subfamily A, 3 (216352_x_at), score: 0.63 PDE5Aphosphodiesterase 5A, cGMP-specific (206757_at), score: 0.73 PHF7PHD finger protein 7 (215622_x_at), score: -0.95 PI4K2Aphosphatidylinositol 4-kinase type 2 alpha (209345_s_at), score: 0.56 PIP5K1Cphosphatidylinositol-4-phosphate 5-kinase, type I, gamma (212518_at), score: 0.77 PKN1protein kinase N1 (202161_at), score: 0.57 PKP2plakophilin 2 (207717_s_at), score: 0.56 PLAUplasminogen activator, urokinase (211668_s_at), score: 0.67 PORP450 (cytochrome) oxidoreductase (208928_at), score: 0.61 PRKCDprotein kinase C, delta (202545_at), score: 0.68 PSMB8proteasome (prosome, macropain) subunit, beta type, 8 (large multifunctional peptidase 7) (209040_s_at), score: 0.56 PTPROprotein tyrosine phosphatase, receptor type, O (211600_at), score: 0.64 PXNpaxillin (211823_s_at), score: 0.71 PYGLphosphorylase, glycogen, liver (202990_at), score: 0.55 RCAN2regulator of calcineurin 2 (203498_at), score: 0.55 REG3Aregenerating islet-derived 3 alpha (205815_at), score: -0.83 RIPK1receptor (TNFRSF)-interacting serine-threonine kinase 1 (209941_at), score: 0.57 RNF130ring finger protein 130 (217865_at), score: 0.62 RNF24ring finger protein 24 (204669_s_at), score: 0.6 RPL21P68ribosomal protein L21 pseudogene 68 (217340_at), score: -0.84 RPS17P5ribosomal protein S17 pseudogene 5 (216348_at), score: -1 SAV1salvador homolog 1 (Drosophila) (218276_s_at), score: 0.71 SCAMP4secretory carrier membrane protein 4 (213244_at), score: 0.73 SDC3syndecan 3 (202898_at), score: 0.56 SDPRserum deprivation response (phosphatidylserine binding protein) (218711_s_at), score: 0.56 SGSHN-sulfoglucosamine sulfohydrolase (35626_at), score: 0.59 SH3GLB2SH3-domain GRB2-like endophilin B2 (218813_s_at), score: 0.63 SHBSrc homology 2 domain containing adaptor protein B (204657_s_at), score: 0.65 SIGIRRsingle immunoglobulin and toll-interleukin 1 receptor (TIR) domain (52940_at), score: 0.62 SIGLEC1sialic acid binding Ig-like lectin 1, sialoadhesin (44673_at), score: -0.83 SIGLEC6sialic acid binding Ig-like lectin 6 (210796_x_at), score: -0.84 SIN3BSIN3 homolog B, transcription regulator (yeast) (209352_s_at), score: 0.62 SLC5A5solute carrier family 5 (sodium iodide symporter), member 5 (211123_at), score: -0.83 SLC9A3R2solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 (209830_s_at), score: 0.65 SMG6Smg-6 homolog, nonsense mediated mRNA decay factor (C. elegans) (214940_s_at), score: 0.58 SNAP25synaptosomal-associated protein, 25kDa (202508_s_at), score: 0.61 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: 0.66 SPARCL1SPARC-like 1 (hevin) (200795_at), score: -0.82 SPATA20spermatogenesis associated 20 (218164_at), score: 0.69 SPRY4sprouty homolog 4 (Drosophila) (221489_s_at), score: 0.62 STOMstomatin (201060_x_at), score: 0.62 SULT1B1sulfotransferase family, cytosolic, 1B, member 1 (207601_at), score: -0.88 SYT1synaptotagmin I (203999_at), score: 0.56 TAPBPTAP binding protein (tapasin) (208829_at), score: 0.7 TBC1D2BTBC1 domain family, member 2B (212796_s_at), score: 0.6 TERF2telomeric repeat binding factor 2 (203611_at), score: 0.6 TGFBR2transforming growth factor, beta receptor II (70/80kDa) (207334_s_at), score: 0.59 TIAF1TGFB1-induced anti-apoptotic factor 1 (202039_at), score: 0.63 TLE6transducin-like enhancer of split 6 (E(sp1) homolog, Drosophila) (222219_s_at), score: -0.85 TNFAIP2tumor necrosis factor, alpha-induced protein 2 (202510_s_at), score: 0.7 TP53I11tumor protein p53 inducible protein 11 (214667_s_at), score: 0.63 TPP1tripeptidyl peptidase I (200742_s_at), score: 0.6 TRIM45tripartite motif-containing 45 (219923_at), score: -0.82 TSC2tuberous sclerosis 2 (215735_s_at), score: 0.63 TSKUtsukushin (218245_at), score: 0.61 UBTD1ubiquitin domain containing 1 (219172_at), score: 0.63 VEGFBvascular endothelial growth factor B (203683_s_at), score: 0.87 VPS37Bvacuolar protein sorting 37 homolog B (S. cerevisiae) (221704_s_at), score: 0.73 WBP2WW domain binding protein 2 (209117_at), score: 0.61 WDR4WD repeat domain 4 (221632_s_at), score: -0.85 WDR42AWD repeat domain 42A (202249_s_at), score: 0.58 WDR6WD repeat domain 6 (217734_s_at), score: 0.63 ZDHHC7zinc finger, DHHC-type containing 7 (218606_at), score: 0.56 ZNF282zinc finger protein 282 (212892_at), score: 0.58
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3407-raw-cel-1437949557.cel | 1 | 4 | Cockayne | hgu133a | CS | eGFP |
| 2Twin.CEL | 2 | 2 | DS-twin | hgu133plus2 | none | DS-twin 2 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 6Twin.CEL | 6 | 2 | DS-twin | hgu133plus2 | none | DS-twin 6 |
| 5CTwin.CEL | 5 | 2 | DS-twin | hgu133plus2 | Down | DS-twin 5 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 1Twin.CEL | 1 | 2 | DS-twin | hgu133plus2 | Down | DS-twin 1 |
| t21d 08-03.CEL | 7 | 1 | DS-CC | hgu133a | Down | DS-CC 7 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| t21c 08-03.CEL | 6 | 1 | DS-CC | hgu133a | Down | DS-CC 6 |
| 4Twin.CEL | 4 | 2 | DS-twin | hgu133plus2 | none | DS-twin 4 |
| E-GEOD-3860-raw-cel-1561690376.cel | 13 | 5 | HGPS | hgu133a | HGPS | AG11513 |
| E-TABM-263-raw-cel-1515486311.cel | 34 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486171.cel | 27 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 46C.CEL | 3 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 3 |
| E-TABM-263-raw-cel-1515486291.cel | 33 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486411.cel | 39 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485791.cel | 8 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485991.cel | 18 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486071.cel | 22 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 46A.CEL | 1 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |