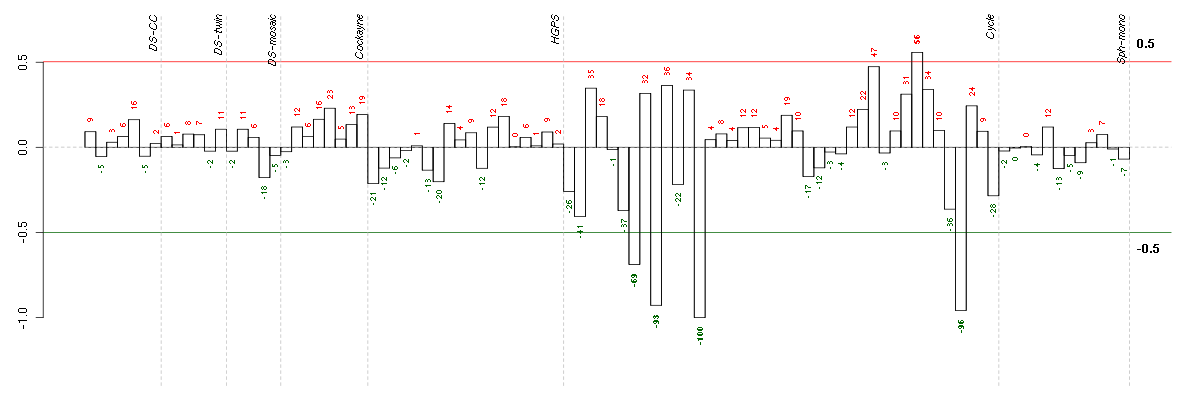

Under-expression is coded with green,
over-expression with red color.

signal transduction
The cascade of processes by which a signal interacts with a receptor, causing a change in the level or activity of a second messenger or other downstream target, and ultimately effecting a change in the functioning of the cell.
cell communication
Any process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
intracellular signaling cascade
A series of reactions within the cell that occur as a result of a single trigger reaction or compound.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
all
This term is the most general term possible
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
signal transduction
The cascade of processes by which a signal interacts with a receptor, causing a change in the level or activity of a second messenger or other downstream target, and ultimately effecting a change in the functioning of the cell.

plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
integral to membrane
Penetrating at least one phospholipid bilayer of a membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer. When used to describe a protein, indicates that all or part of the peptide sequence is embedded in the membrane.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.

calcium ion binding
Interacting selectively with calcium ions (Ca2+).
molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
binding
The selective, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
metal ion binding
Interacting selectively with any metal ion.
ion binding
Interacting selectively with ions, charged atoms or groups of atoms.
cation binding
Interacting selectively with cations, charged atoms or groups of atoms with a net positive charge.
all
This term is the most general term possible
calcium ion binding
Interacting selectively with calcium ions (Ca2+).
| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 04080 | 1.765e-02 | 2.641 | 10 | 78 | Neuroactive ligand-receptor interaction |
ABHD5abhydrolase domain containing 5 (218739_at), score: -0.59 ACN9ACN9 homolog (S. cerevisiae) (218981_at), score: -0.57 ACSL1acyl-CoA synthetase long-chain family member 1 (201963_at), score: -0.62 ACSL4acyl-CoA synthetase long-chain family member 4 (202422_s_at), score: -0.81 ACSS3acyl-CoA synthetase short-chain family member 3 (219616_at), score: 0.54 ACTR3BARP3 actin-related protein 3 homolog B (yeast) (218868_at), score: -0.9 ADARadenosine deaminase, RNA-specific (201786_s_at), score: 0.68 ADH1Balcohol dehydrogenase 1B (class I), beta polypeptide (209612_s_at), score: 0.53 ADNP2ADNP homeobox 2 (203321_s_at), score: 0.54 ADORA2Badenosine A2b receptor (205891_at), score: 0.78 AHI1Abelson helper integration site 1 (221569_at), score: -0.84 AIM1absent in melanoma 1 (212543_at), score: 0.79 AKR1C1aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase) (204151_x_at), score: 0.56 AKR1C3aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II) (209160_at), score: 0.58 ALDH3A2aldehyde dehydrogenase 3 family, member A2 (202053_s_at), score: 0.59 ANGPTL2angiopoietin-like 2 (213004_at), score: 0.6 ANXA10annexin A10 (210143_at), score: -0.81 APAF1apoptotic peptidase activating factor 1 (204859_s_at), score: -0.59 APBA1amyloid beta (A4) precursor protein-binding, family A, member 1 (206679_at), score: -0.59 APOL6apolipoprotein L, 6 (219716_at), score: 0.65 ARFGAP3ADP-ribosylation factor GTPase activating protein 3 (202211_at), score: -0.61 ARHGEF2rho/rac guanine nucleotide exchange factor (GEF) 2 (209435_s_at), score: 0.61 ARSJarylsulfatase family, member J (219973_at), score: -0.6 ASAP3ArfGAP with SH3 domain, ankyrin repeat and PH domain 3 (219103_at), score: 0.54 ATP2C1ATPase, Ca++ transporting, type 2C, member 1 (211137_s_at), score: -0.63 BCAS3breast carcinoma amplified sequence 3 (220488_s_at), score: 0.56 BIN1bridging integrator 1 (210201_x_at), score: 0.54 BMP6bone morphogenetic protein 6 (206176_at), score: -0.88 BRWD1bromodomain and WD repeat domain containing 1 (214820_at), score: -0.58 BTBD2BTB (POZ) domain containing 2 (207722_s_at), score: 0.54 C10orf97chromosome 10 open reading frame 97 (218297_at), score: -0.64 C11orf41chromosome 11 open reading frame 41 (214772_at), score: -0.6 C11orf75chromosome 11 open reading frame 75 (219806_s_at), score: -0.58 C13orf15chromosome 13 open reading frame 15 (218723_s_at), score: 0.64 C14orf45chromosome 14 open reading frame 45 (220173_at), score: -0.68 C14orf94chromosome 14 open reading frame 94 (218383_at), score: 0.59 C15orf33chromosome 15 open reading frame 33 (216411_s_at), score: -0.72 C16orf45chromosome 16 open reading frame 45 (212736_at), score: 0.59 C1orf107chromosome 1 open reading frame 107 (214193_s_at), score: -0.57 C1orf38chromosome 1 open reading frame 38 (210785_s_at), score: 0.64 C4orf43chromosome 4 open reading frame 43 (218513_at), score: -0.8 C5orf44chromosome 5 open reading frame 44 (218674_at), score: -0.71 C7orf64chromosome 7 open reading frame 64 (221596_s_at), score: -0.66 CA9carbonic anhydrase IX (205199_at), score: 0.54 CALB2calbindin 2 (205428_s_at), score: -0.71 CALCOCO1calcium binding and coiled-coil domain 1 (209002_s_at), score: 0.68 CASC3cancer susceptibility candidate 3 (207842_s_at), score: 0.6 CCDC41coiled-coil domain containing 41 (219644_at), score: -0.61 CCDC76coiled-coil domain containing 76 (219130_at), score: -0.59 CDC25Bcell division cycle 25 homolog B (S. pombe) (201853_s_at), score: 0.62 CDC2L6cell division cycle 2-like 6 (CDK8-like) (212899_at), score: 0.63 CDC42EP4CDC42 effector protein (Rho GTPase binding) 4 (218062_x_at), score: 0.64 CDH13cadherin 13, H-cadherin (heart) (204726_at), score: -0.59 CDH6cadherin 6, type 2, K-cadherin (fetal kidney) (205532_s_at), score: -0.72 CEP68centrosomal protein 68kDa (212677_s_at), score: 0.56 CHORDC1cysteine and histidine-rich domain (CHORD)-containing 1 (218566_s_at), score: -0.69 CHUKconserved helix-loop-helix ubiquitous kinase (209666_s_at), score: -0.67 CICcapicua homolog (Drosophila) (212784_at), score: 0.59 CMPK1cytidine monophosphate (UMP-CMP) kinase 1, cytosolic (217870_s_at), score: -0.61 CRBNcereblon (218142_s_at), score: 0.62 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: 0.97 CSGALNACT2chondroitin sulfate N-acetylgalactosaminyltransferase 2 (222235_s_at), score: -0.62 CTAGE5CTAGE family, member 5 (215930_s_at), score: -0.69 CTDSP2CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 (203445_s_at), score: 0.53 CXCL5chemokine (C-X-C motif) ligand 5 (214974_x_at), score: -0.58 CXCL6chemokine (C-X-C motif) ligand 6 (granulocyte chemotactic protein 2) (206336_at), score: -0.6 DCLK1doublecortin-like kinase 1 (205399_at), score: 0.65 DDAH2dimethylarginine dimethylaminohydrolase 2 (215537_x_at), score: 0.54 DDX52DEAD (Asp-Glu-Ala-Asp) box polypeptide 52 (212834_at), score: -0.6 DENND2ADENN/MADD domain containing 2A (53991_at), score: 0.68 DENND5BDENN/MADD domain containing 5B (215058_at), score: -0.81 DEPDC6DEP domain containing 6 (218858_at), score: 0.66 DHRS3dehydrogenase/reductase (SDR family) member 3 (202481_at), score: 0.71 DIS3DIS3 mitotic control homolog (S. cerevisiae) (218362_s_at), score: -0.65 DNAJB9DnaJ (Hsp40) homolog, subfamily B, member 9 (202843_at), score: -0.88 DNAJC6DnaJ (Hsp40) homolog, subfamily C, member 6 (204720_s_at), score: -0.67 DNM1dynamin 1 (215116_s_at), score: 0.55 DOCK10dedicator of cytokinesis 10 (219279_at), score: -0.71 DOPEY1dopey family member 1 (40612_at), score: -0.61 DPF3D4, zinc and double PHD fingers, family 3 (219746_at), score: -0.61 DUSP4dual specificity phosphatase 4 (204014_at), score: -0.77 EAF2ELL associated factor 2 (219551_at), score: -0.95 EDEM1ER degradation enhancer, mannosidase alpha-like 1 (203279_at), score: -0.67 EGFepidermal growth factor (beta-urogastrone) (206254_at), score: -0.8 EIF2AK3eukaryotic translation initiation factor 2-alpha kinase 3 (218696_at), score: -0.64 EIF4Eeukaryotic translation initiation factor 4E (201436_at), score: -0.85 EIF5Beukaryotic translation initiation factor 5B (214314_s_at), score: -0.57 ENTPD1ectonucleoside triphosphate diphosphohydrolase 1 (209473_at), score: -0.63 ESM1endothelial cell-specific molecule 1 (208394_x_at), score: -0.82 EVI1ecotropic viral integration site 1 (221884_at), score: -0.61 F2RL2coagulation factor II (thrombin) receptor-like 2 (206795_at), score: -0.59 FAM102Afamily with sequence similarity 102, member A (212400_at), score: 0.52 FAM155Afamily with sequence similarity 155, member A (214825_at), score: -0.7 FAM18Bfamily with sequence similarity 18, member B (218446_s_at), score: -0.7 FAM8A1family with sequence similarity 8, member A1 (203420_at), score: -0.61 FBXL14F-box and leucine-rich repeat protein 14 (213145_at), score: 0.68 FERMT1fermitin family homolog 1 (Drosophila) (218796_at), score: -0.74 FICDFIC domain containing (219910_at), score: -0.73 FLJ12529pre-mRNA cleavage factor I, 59 kDa subunit (217866_at), score: 0.57 FNBP1formin binding protein 1 (212288_at), score: 0.56 FNDC3Afibronectin type III domain containing 3A (202304_at), score: -0.61 FOXK2forkhead box K2 (203064_s_at), score: 0.56 FSTL3follistatin-like 3 (secreted glycoprotein) (203592_s_at), score: -0.59 FUBP3far upstream element (FUSE) binding protein 3 (212824_at), score: -0.57 FYNFYN oncogene related to SRC, FGR, YES (210105_s_at), score: 0.53 G3BP2GTPase activating protein (SH3 domain) binding protein 2 (208841_s_at), score: -0.62 GALK2galactokinase 2 (205219_s_at), score: -0.68 GAS7growth arrest-specific 7 (202191_s_at), score: 0.53 GCS1glucosidase I (210627_s_at), score: -0.58 GDF15growth differentiation factor 15 (221577_x_at), score: 0.54 GFOD1glucose-fructose oxidoreductase domain containing 1 (219821_s_at), score: -0.89 GHRgrowth hormone receptor (205498_at), score: -0.85 GKglycerol kinase (207387_s_at), score: -0.86 GK3Pglycerol kinase 3 pseudogene (215966_x_at), score: -0.98 GMCL1germ cell-less homolog 1 (Drosophila) (218458_at), score: -0.9 GMPRguanosine monophosphate reductase (204187_at), score: 0.6 GOLT1Bgolgi transport 1 homolog B (S. cerevisiae) (218193_s_at), score: -0.66 GPR177G protein-coupled receptor 177 (221958_s_at), score: -0.62 GPRC5BG protein-coupled receptor, family C, group 5, member B (203632_s_at), score: 0.57 GPSM2G-protein signaling modulator 2 (AGS3-like, C. elegans) (221922_at), score: 0.53 GRB14growth factor receptor-bound protein 14 (206204_at), score: -0.61 GTF2A1Lgeneral transcription factor IIA, 1-like (213413_at), score: 0.73 H1FXH1 histone family, member X (204805_s_at), score: 0.67 HDGFhepatoma-derived growth factor (high-mobility group protein 1-like) (200896_x_at), score: 0.68 HISPPD1histidine acid phosphatase domain containing 1 (203253_s_at), score: -0.78 HOXC5homeobox C5 (206739_at), score: -0.67 HRH1histamine receptor H1 (205580_s_at), score: 0.53 HSD17B6hydroxysteroid (17-beta) dehydrogenase 6 homolog (mouse) (37512_at), score: -0.66 HSD17B7hydroxysteroid (17-beta) dehydrogenase 7 (220081_x_at), score: 0.56 HSPA12Aheat shock 70kDa protein 12A (214434_at), score: 0.71 HTR2A5-hydroxytryptamine (serotonin) receptor 2A (207135_at), score: -0.57 ID4inhibitor of DNA binding 4, dominant negative helix-loop-helix protein (209291_at), score: -0.6 IFIH1interferon induced with helicase C domain 1 (219209_at), score: 0.59 IL33interleukin 33 (209821_at), score: -0.77 IRF1interferon regulatory factor 1 (202531_at), score: 0.59 IRS1insulin receptor substrate 1 (204686_at), score: -0.64 ITGA1integrin, alpha 1 (214660_at), score: -0.63 ITGAVintegrin, alpha V (vitronectin receptor, alpha polypeptide, antigen CD51) (202351_at), score: -0.62 ITPR2inositol 1,4,5-triphosphate receptor, type 2 (202660_at), score: -0.65 JUNDjun D proto-oncogene (203751_x_at), score: 0.6 JUPjunction plakoglobin (201015_s_at), score: 0.59 KAL1Kallmann syndrome 1 sequence (205206_at), score: -0.99 KCNJ8potassium inwardly-rectifying channel, subfamily J, member 8 (205303_at), score: -0.64 KIAA0562KIAA0562 (204075_s_at), score: -0.68 KIAA1305KIAA1305 (220911_s_at), score: 0.62 KIAA1462KIAA1462 (213316_at), score: 0.69 KLHL21kelch-like 21 (Drosophila) (203068_at), score: -0.58 KLHL23kelch-like 23 (Drosophila) (213610_s_at), score: -0.69 LARP4La ribonucleoprotein domain family, member 4 (214155_s_at), score: -0.83 LDLRlow density lipoprotein receptor (202068_s_at), score: 0.55 LHFPL2lipoma HMGIC fusion partner-like 2 (212658_at), score: 0.59 LIG4ligase IV, DNA, ATP-dependent (206235_at), score: -0.58 LIN7Blin-7 homolog B (C. elegans) (219760_at), score: -0.73 LOC100132540similar to LOC339047 protein (214870_x_at), score: 0.81 LOC339047hypothetical protein LOC339047 (221501_x_at), score: 0.75 LOC391132similar to hCG2041276 (216177_at), score: -0.63 LOC399491LOC399491 protein (214035_x_at), score: 0.76 LRDDleucine-rich repeats and death domain containing (221640_s_at), score: 0.55 LRP8low density lipoprotein receptor-related protein 8, apolipoprotein e receptor (205282_at), score: -0.67 LRRN3leucine rich repeat neuronal 3 (209841_s_at), score: -0.8 LRRTM4leucine rich repeat transmembrane neuronal 4 (220345_at), score: 0.53 LYPD1LY6/PLAUR domain containing 1 (212909_at), score: -0.78 MAN1C1mannosidase, alpha, class 1C, member 1 (218918_at), score: 0.59 MAP2K5mitogen-activated protein kinase kinase 5 (211370_s_at), score: 0.57 MBOAT2membrane bound O-acyltransferase domain containing 2 (213288_at), score: -0.57 MC4Rmelanocortin 4 receptor (221467_at), score: -0.67 MFAP3microfibrillar-associated protein 3 (213123_at), score: -0.57 MFAP3Lmicrofibrillar-associated protein 3-like (205442_at), score: -0.84 MGC87042similar to Six transmembrane epithelial antigen of prostate (217553_at), score: -0.59 MKNK2MAP kinase interacting serine/threonine kinase 2 (218205_s_at), score: 0.59 MMP10matrix metallopeptidase 10 (stromelysin 2) (205680_at), score: -0.72 MMP16matrix metallopeptidase 16 (membrane-inserted) (207012_at), score: -0.69 MSTO1misato homolog 1 (Drosophila) (218296_x_at), score: -0.76 MYBBP1AMYB binding protein (P160) 1a (219098_at), score: -0.59 NCALDneurocalcin delta (211685_s_at), score: -0.8 NEDD4neural precursor cell expressed, developmentally down-regulated 4 (213012_at), score: -0.63 NEFMneurofilament, medium polypeptide (205113_at), score: -0.97 NEO1neogenin homolog 1 (chicken) (204321_at), score: 0.54 NMIN-myc (and STAT) interactor (203964_at), score: 0.65 NOX4NADPH oxidase 4 (219773_at), score: -0.76 NPIPnuclear pore complex interacting protein (204538_x_at), score: 0.83 NPR2natriuretic peptide receptor B/guanylate cyclase B (atrionatriuretic peptide receptor B) (204310_s_at), score: 0.54 NPTX1neuronal pentraxin I (204684_at), score: -0.82 NR5A2nuclear receptor subfamily 5, group A, member 2 (208343_s_at), score: -0.58 NRXN3neurexin 3 (205795_at), score: -0.69 NTMneurotrimin (222020_s_at), score: -0.59 NUP160nucleoporin 160kDa (212709_at), score: -0.6 OAS22'-5'-oligoadenylate synthetase 2, 69/71kDa (204972_at), score: 0.56 OBFC2Aoligonucleotide/oligosaccharide-binding fold containing 2A (219334_s_at), score: -0.67 OLR1oxidized low density lipoprotein (lectin-like) receptor 1 (210004_at), score: -0.63 OPCMLopioid binding protein/cell adhesion molecule-like (214111_at), score: -0.76 ORAI3ORAI calcium release-activated calcium modulator 3 (221864_at), score: 0.55 OSGIN2oxidative stress induced growth inhibitor family member 2 (204024_at), score: -0.58 OSTM1osteopetrosis associated transmembrane protein 1 (218196_at), score: -0.71 P2RY5purinergic receptor P2Y, G-protein coupled, 5 (218589_at), score: 0.67 PAQR3progestin and adipoQ receptor family member III (213372_at), score: -0.71 PCDH9protocadherin 9 (219737_s_at), score: -1 PCYT2phosphate cytidylyltransferase 2, ethanolamine (209577_at), score: 0.55 PDCD1LG2programmed cell death 1 ligand 2 (220049_s_at), score: -0.61 PDE5Aphosphodiesterase 5A, cGMP-specific (206757_at), score: 0.55 PHF16PHD finger protein 16 (204866_at), score: -0.6 PIONpigeon homolog (Drosophila) (222150_s_at), score: -0.69 PITX1paired-like homeodomain 1 (208502_s_at), score: 0.63 PKIGprotein kinase (cAMP-dependent, catalytic) inhibitor gamma (202732_at), score: 0.56 PLAUplasminogen activator, urokinase (211668_s_at), score: 0.53 PLCD1phospholipase C, delta 1 (205125_at), score: 0.52 PMP22peripheral myelin protein 22 (210139_s_at), score: 0.71 PNPLA2patatin-like phospholipase domain containing 2 (212705_x_at), score: 0.7 PPARGperoxisome proliferator-activated receptor gamma (208510_s_at), score: 0.63 PPP3R1protein phosphatase 3 (formerly 2B), regulatory subunit B, alpha isoform (204507_s_at), score: -0.73 PRKCHprotein kinase C, eta (218764_at), score: 0.55 PRMT3protein arginine methyltransferase 3 (213320_at), score: -0.69 PRSS2protease, serine, 2 (trypsin 2) (205402_x_at), score: -0.57 PTHLHparathyroid hormone-like hormone (211756_at), score: -0.64 PTP4A1protein tyrosine phosphatase type IVA, member 1 (200730_s_at), score: -0.66 PTPROprotein tyrosine phosphatase, receptor type, O (211600_at), score: 0.55 RAB1ARAB1A, member RAS oncogene family (213440_at), score: -0.7 RAB7ARAB7A, member RAS oncogene family (211960_s_at), score: -0.67 RANBP6RAN binding protein 6 (213019_at), score: -0.6 RASSF2Ras association (RalGDS/AF-6) domain family member 2 (203185_at), score: 0.62 RBM15BRNA binding motif protein 15B (202689_at), score: 0.53 RBM47RNA binding motif protein 47 (218035_s_at), score: 0.54 REREarginine-glutamic acid dipeptide (RE) repeats (200940_s_at), score: 0.71 RFX5regulatory factor X, 5 (influences HLA class II expression) (202963_at), score: 0.55 RINT1RAD50 interactor 1 (218598_at), score: -0.6 RNF11ring finger protein 11 (208924_at), score: -0.66 RNF220ring finger protein 220 (219988_s_at), score: 0.65 RNF44ring finger protein 44 (203286_at), score: 0.62 RNFT1ring finger protein, transmembrane 1 (221194_s_at), score: -0.6 ROR2receptor tyrosine kinase-like orphan receptor 2 (205578_at), score: 0.64 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: -0.6 RPL23ribosomal protein L23 (214744_s_at), score: -0.6 RPLP2P1ribosomal protein, large P2, pseudogene 1 (216490_x_at), score: -0.59 RSC1A1regulatory solute carrier protein, family 1, member 1 (214583_at), score: -0.58 S1PR1sphingosine-1-phosphate receptor 1 (204642_at), score: -0.58 SAP30LSAP30-like (219129_s_at), score: 0.65 SAV1salvador homolog 1 (Drosophila) (218276_s_at), score: 0.55 SBNO2strawberry notch homolog 2 (Drosophila) (204166_at), score: 0.53 SCAMP4secretory carrier membrane protein 4 (213244_at), score: 0.59 SDF2L1stromal cell-derived factor 2-like 1 (218681_s_at), score: -0.65 SERPINB2serpin peptidase inhibitor, clade B (ovalbumin), member 2 (204614_at), score: -0.63 SERPINB9serpin peptidase inhibitor, clade B (ovalbumin), member 9 (209723_at), score: 0.62 SF1splicing factor 1 (208313_s_at), score: 0.58 SH3BP5SH3-domain binding protein 5 (BTK-associated) (201810_s_at), score: 0.55 SIGIRRsingle immunoglobulin and toll-interleukin 1 receptor (TIR) domain (52940_at), score: 0.61 SIX1SIX homeobox 1 (205817_at), score: 0.54 SLC25A13solute carrier family 25, member 13 (citrin) (203775_at), score: -0.67 SLC25A32solute carrier family 25, member 32 (221020_s_at), score: -0.6 SLC2A6solute carrier family 2 (facilitated glucose transporter), member 6 (220091_at), score: 0.56 SLC33A1solute carrier family 33 (acetyl-CoA transporter), member 1 (203165_s_at), score: -0.83 SLC39A7solute carrier family 39 (zinc transporter), member 7 (202667_s_at), score: -0.58 SLC4A4solute carrier family 4, sodium bicarbonate cotransporter, member 4 (203908_at), score: -0.65 SLC4A7solute carrier family 4, sodium bicarbonate cotransporter, member 7 (209884_s_at), score: -0.88 SLC6A9solute carrier family 6 (neurotransmitter transporter, glycine), member 9 (207043_s_at), score: 0.55 SLC7A6solute carrier family 7 (cationic amino acid transporter, y+ system), member 6 (203578_s_at), score: -0.58 SLC8A1solute carrier family 8 (sodium/calcium exchanger), member 1 (207053_at), score: -0.69 SLMO2slowmo homolog 2 (Drosophila) (217851_s_at), score: -0.81 SMAD3SMAD family member 3 (218284_at), score: 0.7 SMN1survival of motor neuron 1, telomeric (203852_s_at), score: -0.63 SNNstannin (218032_at), score: 0.8 SPRY1sprouty homolog 1, antagonist of FGF signaling (Drosophila) (212558_at), score: 0.56 SSTR1somatostatin receptor 1 (208482_at), score: -0.81 ST20suppressor of tumorigenicity 20 (210242_x_at), score: -0.59 STIM1stromal interaction molecule 1 (202764_at), score: 0.55 SYNE1spectrin repeat containing, nuclear envelope 1 (209447_at), score: -0.65 SYNE2spectrin repeat containing, nuclear envelope 2 (202761_s_at), score: 0.54 SYNJ1synaptojanin 1 (212990_at), score: -0.65 TAF13TAF13 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 18kDa (205966_at), score: -0.76 TAF1ATATA box binding protein (TBP)-associated factor, RNA polymerase I, A, 48kDa (206613_s_at), score: -0.61 TBC1D2BTBC1 domain family, member 2B (212796_s_at), score: 0.59 TBC1D8TBC1 domain family, member 8 (with GRAM domain) (204526_s_at), score: 0.61 TBC1D8BTBC1 domain family, member 8B (with GRAM domain) (219771_at), score: -0.63 TFDP2transcription factor Dp-2 (E2F dimerization partner 2) (203588_s_at), score: 0.64 TGDSTDP-glucose 4,6-dehydratase (208249_s_at), score: -0.76 THSD7Athrombospondin, type I, domain containing 7A (214920_at), score: 0.66 TICAM2toll-like receptor adaptor molecule 2 (214658_at), score: -0.74 TLR3toll-like receptor 3 (206271_at), score: 0.67 TM4SF1transmembrane 4 L six family member 1 (215034_s_at), score: -0.63 TM9SF3transmembrane 9 superfamily member 3 (217758_s_at), score: -0.57 TMED7transmembrane emp24 protein transport domain containing 7 (209404_s_at), score: -0.57 TMEFF1transmembrane protein with EGF-like and two follistatin-like domains 1 (205122_at), score: -0.68 TMEM2transmembrane protein 2 (218113_at), score: -0.75 TMEM30Atransmembrane protein 30A (217743_s_at), score: -0.65 TMF1TATA element modulatory factor 1 (213024_at), score: -0.64 TNFAIP6tumor necrosis factor, alpha-induced protein 6 (206026_s_at), score: 0.62 TNFSF15tumor necrosis factor (ligand) superfamily, member 15 (221085_at), score: 0.62 TNS3tensin 3 (217853_at), score: 0.56 TOMM20translocase of outer mitochondrial membrane 20 homolog (yeast) (200662_s_at), score: -0.62 TP53tumor protein p53 (201746_at), score: 0.74 TPBGtrophoblast glycoprotein (203476_at), score: 0.59 TRIM21tripartite motif-containing 21 (204804_at), score: 0.73 TRIM23tripartite motif-containing 23 (204732_s_at), score: -0.57 TRIM24tripartite motif-containing 24 (213301_x_at), score: -0.58 TRMT2ATRM2 tRNA methyltransferase 2 homolog A (S. cerevisiae) (91617_at), score: -0.57 TRPC4transient receptor potential cation channel, subfamily C, member 4 (220817_at), score: -0.69 TTLL5tubulin tyrosine ligase-like family, member 5 (214672_at), score: 0.53 UBE2Wubiquitin-conjugating enzyme E2W (putative) (218521_s_at), score: -0.59 UFM1ubiquitin-fold modifier 1 (218050_at), score: -0.63 UGDHUDP-glucose dehydrogenase (203343_at), score: -0.62 UHRF1BP1LUHRF1 binding protein 1-like (213118_at), score: -0.63 ULBP2UL16 binding protein 2 (221291_at), score: -0.69 ULK1unc-51-like kinase 1 (C. elegans) (209333_at), score: 0.61 URB1URB1 ribosome biogenesis 1 homolog (S. cerevisiae) (212996_s_at), score: -0.82 VWA5Avon Willebrand factor A domain containing 5A (210102_at), score: 0.58 WBP2WW domain binding protein 2 (209117_at), score: 0.54 WDR4WD repeat domain 4 (221632_s_at), score: -0.73 WDR6WD repeat domain 6 (217734_s_at), score: 0.63 WHAMML1WAS protein homolog associated with actin, golgi membranes and microtubules-like 1 (213908_at), score: -0.57 YDD19YDD19 protein (37079_at), score: -0.65 YES1v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 (202932_at), score: 0.59 ZBTB20zinc finger and BTB domain containing 20 (205383_s_at), score: 0.53 ZBTB6zinc finger and BTB domain containing 6 (206098_at), score: -0.67 ZCCHC10zinc finger, CCHC domain containing 10 (221193_s_at), score: -0.59 ZDHHC7zinc finger, DHHC-type containing 7 (218606_at), score: 0.89 ZFAND1zinc finger, AN1-type domain 1 (218919_at), score: -0.62 ZNF23zinc finger protein 23 (KOX 16) (213934_s_at), score: -0.66 ZNF248zinc finger protein 248 (213269_at), score: -0.57
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486291.cel | 33 | 6 | Cycle | hgu133a2 | none | Cycle 1 |