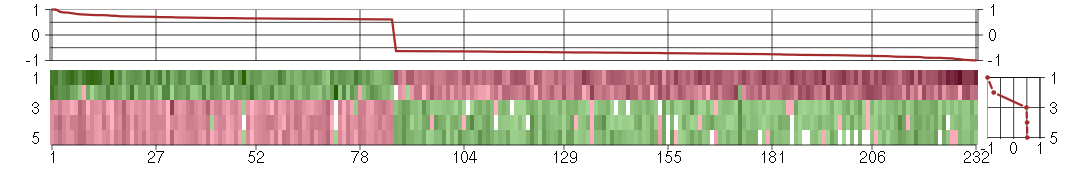

Under-expression is coded with green,
over-expression with red color.



ABAT4-aminobutyrate aminotransferase (209459_s_at), score: 0.64 ABCB1ATP-binding cassette, sub-family B (MDR/TAP), member 1 (209993_at), score: -0.91 ABLIM1actin binding LIM protein 1 (200965_s_at), score: 0.67 ACSL3acyl-CoA synthetase long-chain family member 3 (201660_at), score: 0.69 ACTR8ARP8 actin-related protein 8 homolog (yeast) (218658_s_at), score: 0.78 ADAMTS1ADAM metallopeptidase with thrombospondin type 1 motif, 1 (222162_s_at), score: 0.68 AFF4AF4/FMR2 family, member 4 (219199_at), score: -0.69 AKAP10A kinase (PRKA) anchor protein 10 (205045_at), score: -0.74 AKAP12A kinase (PRKA) anchor protein 12 (210517_s_at), score: 0.66 AKAP8LA kinase (PRKA) anchor protein 8-like (218064_s_at), score: -0.65 ARF5ADP-ribosylation factor 5 (201526_at), score: -0.67 ARHGAP29Rho GTPase activating protein 29 (203910_at), score: 0.79 ARSJarylsulfatase family, member J (219973_at), score: 0.63 ATP11AATPase, class VI, type 11A (215842_s_at), score: -0.79 ATP6V1AATPase, H+ transporting, lysosomal 70kDa, V1 subunit A (201971_s_at), score: -0.78 ATP9AATPase, class II, type 9A (212062_at), score: 0.67 BAZ2Abromodomain adjacent to zinc finger domain, 2A (201353_s_at), score: -0.71 BET1Lblocked early in transport 1 homolog (S. cerevisiae)-like (220470_at), score: -0.68 C10orf116chromosome 10 open reading frame 116 (203571_s_at), score: 0.91 C15orf29chromosome 15 open reading frame 29 (218791_s_at), score: 0.64 C20orf149chromosome 20 open reading frame 149 (218010_x_at), score: 0.72 C3orf60chromosome 3 open reading frame 60 (209177_at), score: 0.61 C4orf30chromosome 4 open reading frame 30 (219717_at), score: -0.86 C5orf30chromosome 5 open reading frame 30 (221823_at), score: 0.68 CAMSAP1L1calmodulin regulated spectrin-associated protein 1-like 1 (212763_at), score: -0.65 CATSPER2cation channel, sperm associated 2 (217588_at), score: -0.65 CD44CD44 molecule (Indian blood group) (210916_s_at), score: -0.71 CDK5R1cyclin-dependent kinase 5, regulatory subunit 1 (p35) (204995_at), score: 0.64 CHMchoroideremia (Rab escort protein 1) (207099_s_at), score: -0.8 CLINT1clathrin interactor 1 (201768_s_at), score: -0.77 CLN3ceroid-lipofuscinosis, neuronal 3 (209275_s_at), score: -0.86 CNNM3cyclin M3 (220739_s_at), score: -0.73 COL13A1collagen, type XIII, alpha 1 (211343_s_at), score: 0.7 COL18A1collagen, type XVIII, alpha 1 (209081_s_at), score: -0.75 COPAcoatomer protein complex, subunit alpha (214336_s_at), score: -0.71 COPS7ACOP9 constitutive photomorphogenic homolog subunit 7A (Arabidopsis) (209029_at), score: -0.7 CPDcarboxypeptidase D (201942_s_at), score: -0.83 CREBL2cAMP responsive element binding protein-like 2 (201990_s_at), score: 0.72 CRLF3cytokine receptor-like factor 3 (205474_at), score: 0.65 CRYBG3beta-gamma crystallin domain containing 3 (214030_at), score: 0.64 CSNK1A1casein kinase 1, alpha 1 (206562_s_at), score: -0.64 CTNND1catenin (cadherin-associated protein), delta 1 (208407_s_at), score: 0.61 CTTNcortactin (201059_at), score: 0.65 CUL3cullin 3 (201370_s_at), score: 0.62 DAZAP2DAZ associated protein 2 (212595_s_at), score: -0.82 DDX17DEAD (Asp-Glu-Ala-Asp) box polypeptide 17 (208151_x_at), score: -0.81 DDX3XDEAD (Asp-Glu-Ala-Asp) box polypeptide 3, X-linked (201211_s_at), score: -0.71 DHDDSdehydrodolichyl diphosphate synthase (218547_at), score: 0.73 DHX9DEAH (Asp-Glu-Ala-His) box polypeptide 9 (212105_s_at), score: -0.97 DKK1dickkopf homolog 1 (Xenopus laevis) (204602_at), score: 0.71 DMWDdystrophia myotonica, WD repeat containing (33768_at), score: -0.79 DNAJB1DnaJ (Hsp40) homolog, subfamily B, member 1 (200664_s_at), score: -0.84 DNAJC3DnaJ (Hsp40) homolog, subfamily C, member 3 (208499_s_at), score: -0.9 DND1dead end homolog 1 (zebrafish) (57739_at), score: -0.82 DRAMdamage-regulated autophagy modulator (218627_at), score: 0.68 DTX3deltex homolog 3 (Drosophila) (49049_at), score: -0.77 DUSP10dual specificity phosphatase 10 (215501_s_at), score: -0.79 DUSP5dual specificity phosphatase 5 (209457_at), score: 0.65 ENOSF1enolase superfamily member 1 (204142_at), score: 0.64 EPHB3EPH receptor B3 (1438_at), score: -0.66 EPHX1epoxide hydrolase 1, microsomal (xenobiotic) (202017_at), score: -0.63 ERGIC2ERGIC and golgi 2 (218135_at), score: 0.62 ETS1v-ets erythroblastosis virus E26 oncogene homolog 1 (avian) (214447_at), score: -0.79 FAM169Afamily with sequence similarity 169, member A (213954_at), score: -0.78 FAM46Cfamily with sequence similarity 46, member C (220306_at), score: 0.7 FAM65Bfamily with sequence similarity 65, member B (209829_at), score: 0.63 FBN2fibrillin 2 (203184_at), score: 0.75 FCARFc fragment of IgA, receptor for (211305_x_at), score: -0.64 FGF12fibroblast growth factor 12 (207501_s_at), score: -0.64 FILIP1Lfilamin A interacting protein 1-like (204135_at), score: 0.63 FLJ12529pre-mRNA cleavage factor I, 59 kDa subunit (217866_at), score: -0.66 FLRT1fibronectin leucine rich transmembrane protein 1 (210414_at), score: -0.68 FOXL2forkhead box L2 (220102_at), score: -0.65 FXR1fragile X mental retardation, autosomal homolog 1 (201635_s_at), score: -0.69 GABPAGA binding protein transcription factor, alpha subunit 60kDa (210188_at), score: -0.81 GABREgamma-aminobutyric acid (GABA) A receptor, epsilon (204537_s_at), score: -0.99 GABRQgamma-aminobutyric acid (GABA) receptor, theta (220886_at), score: -0.64 GAGE4G antigen 4 (207086_x_at), score: -0.69 GAGE6G antigen 6 (208155_x_at), score: -0.64 GATA3GATA binding protein 3 (209604_s_at), score: -0.69 GDE1glycerophosphodiester phosphodiesterase 1 (202593_s_at), score: -0.75 GFRA1GDNF family receptor alpha 1 (205696_s_at), score: 0.73 GLE1GLE1 RNA export mediator homolog (yeast) (206920_s_at), score: -0.69 GLT25D2glycosyltransferase 25 domain containing 2 (209883_at), score: 0.61 GLTPglycolipid transfer protein (219267_at), score: 0.63 GNA12guanine nucleotide binding protein (G protein) alpha 12 (221737_at), score: 0.85 GNA13guanine nucleotide binding protein (G protein), alpha 13 (206917_at), score: -0.78 GNSglucosamine (N-acetyl)-6-sulfatase (203676_at), score: -0.69 GPR137G protein-coupled receptor 137 (43934_at), score: -0.91 GRK5G protein-coupled receptor kinase 5 (204396_s_at), score: 0.66 GTDC1glycosyltransferase-like domain containing 1 (219770_at), score: 0.61 GTF2Igeneral transcription factor II, i (210892_s_at), score: -0.75 H2AFYH2A histone family, member Y (214500_at), score: -0.64 HAGHhydroxyacylglutathione hydrolase (205012_s_at), score: 0.64 HERC5hect domain and RLD 5 (219863_at), score: -0.67 HIPK1homeodomain interacting protein kinase 1 (212291_at), score: -0.81 HOXA10homeobox A10 (213150_at), score: 0.7 HOXB5homeobox B5 (205601_s_at), score: -0.64 HP1BP3heterochromatin protein 1, binding protein 3 (220633_s_at), score: -0.74 HSPA2heat shock 70kDa protein 2 (211538_s_at), score: 0.65 IGF2BP2insulin-like growth factor 2 mRNA binding protein 2 (218847_at), score: 0.69 IKZF5IKAROS family zinc finger 5 (Pegasus) (220086_at), score: -0.75 IPO8importin 8 (205701_at), score: -0.98 ITGB5integrin, beta 5 (214021_x_at), score: 0.72 KIAA0467KIAA0467 (203900_at), score: -0.7 KIF3Bkinesin family member 3B (203943_at), score: -0.9 KPNA4karyopherin alpha 4 (importin alpha 3) (209653_at), score: -0.72 KRT19keratin 19 (201650_at), score: 0.67 KYNUkynureninase (L-kynurenine hydrolase) (217388_s_at), score: -0.74 LOC100170939glucuronidase, beta pseudogene (214850_at), score: -0.65 LOC643313similar to hypothetical protein LOC284701 (211050_x_at), score: -0.66 LOC727977hypothetical LOC727977 (205787_x_at), score: -0.8 LOC730092RRN3 RNA polymerase I transcription factor homolog (S. cerevisiae) pseudogene (216908_x_at), score: -0.71 LRIG2leucine-rich repeats and immunoglobulin-like domains 2 (205953_at), score: -0.74 LRRC2leucine rich repeat containing 2 (219949_at), score: 1 MAFBv-maf musculoaponeurotic fibrosarcoma oncogene homolog B (avian) (218559_s_at), score: -0.86 MAFFv-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian) (36711_at), score: 0.63 MAP2K1mitogen-activated protein kinase kinase 1 (202670_at), score: 0.65 MAP3K2mitogen-activated protein kinase kinase kinase 2 (221695_s_at), score: -0.79 MAP3K7mitogen-activated protein kinase kinase kinase 7 (211537_x_at), score: -0.7 MAP3K7IP2mitogen-activated protein kinase kinase kinase 7 interacting protein 2 (210284_s_at), score: -0.79 MARCH2membrane-associated ring finger (C3HC4) 2 (210075_at), score: 0.65 MAZMYC-associated zinc finger protein (purine-binding transcription factor) (207824_s_at), score: -0.76 MBNL2muscleblind-like 2 (Drosophila) (205018_s_at), score: -0.74 MCRS1microspherule protein 1 (202556_s_at), score: -0.66 MFAP5microfibrillar associated protein 5 (213764_s_at), score: 0.66 MMP1matrix metallopeptidase 1 (interstitial collagenase) (204475_at), score: 0.63 MPDU1mannose-P-dolichol utilization defect 1 (209208_at), score: -0.63 MTMR1myotubularin related protein 1 (213511_s_at), score: 0.62 MTMR12myotubularin related protein 12 (220953_s_at), score: -0.73 MYO10myosin X (201976_s_at), score: 0.64 NBPF14neuroblastoma breakpoint family, member 14 (201104_x_at), score: -0.66 NCOA2nuclear receptor coactivator 2 (205732_s_at), score: -0.73 NCRNA00084non-protein coding RNA 84 (214657_s_at), score: -0.78 NF1neurofibromin 1 (211094_s_at), score: -0.66 NONOnon-POU domain containing, octamer-binding (208698_s_at), score: -0.85 NOTCH2Notch homolog 2 (Drosophila) (210756_s_at), score: -0.77 NRBP1nuclear receptor binding protein 1 (217765_at), score: -0.78 NRP2neuropilin 2 (214632_at), score: -0.75 NT5E5'-nucleotidase, ecto (CD73) (203939_at), score: 0.62 NXF1nuclear RNA export factor 1 (208922_s_at), score: -0.7 OASL2'-5'-oligoadenylate synthetase-like (210797_s_at), score: -1 OGDHoxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) (201282_at), score: -0.67 OXA1Loxidase (cytochrome c) assembly 1-like (208717_at), score: -0.83 OXTRoxytocin receptor (206825_at), score: 0.8 PBLDphenazine biosynthesis-like protein domain containing (219543_at), score: 0.8 PCDHGA3protocadherin gamma subfamily A, 3 (216352_x_at), score: -0.66 PCIF1PDX1 C-terminal inhibiting factor 1 (222045_s_at), score: -0.89 PDPRpyruvate dehydrogenase phosphatase regulatory subunit (220236_at), score: -0.76 PGRMC2progesterone receptor membrane component 2 (213227_at), score: 0.78 PILRBpaired immunoglobin-like type 2 receptor beta (220954_s_at), score: -0.8 POLDIP3polymerase (DNA-directed), delta interacting protein 3 (215357_s_at), score: -0.72 POM121POM121 membrane glycoprotein (rat) (212178_s_at), score: -0.63 POM121L9PPOM121 membrane glycoprotein-like 9 (rat) pseudogene (222253_s_at), score: -0.63 PPP1R3Dprotein phosphatase 1, regulatory (inhibitor) subunit 3D (204554_at), score: 0.68 PPP3CBprotein phosphatase 3 (formerly 2B), catalytic subunit, beta isoform (209817_at), score: 0.65 PRCCpapillary renal cell carcinoma (translocation-associated) (208938_at), score: -0.89 PRKD3protein kinase D3 (211084_x_at), score: -0.86 PRPF40APRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) (214941_s_at), score: -0.73 PSMB4proteasome (prosome, macropain) subunit, beta type, 4 (202243_s_at), score: 0.75 PTX3pentraxin-related gene, rapidly induced by IL-1 beta (206157_at), score: 0.61 PYROXD1pyridine nucleotide-disulphide oxidoreductase domain 1 (213878_at), score: 0.64 RAB27BRAB27B, member RAS oncogene family (207018_s_at), score: -0.63 RAB5CRAB5C, member RAS oncogene family (201156_s_at), score: -0.73 RAB7L1RAB7, member RAS oncogene family-like 1 (218700_s_at), score: -0.69 RABGAP1LRAB GTPase activating protein 1-like (213982_s_at), score: 0.78 RAD23BRAD23 homolog B (S. cerevisiae) (201223_s_at), score: 1 RBM3RNA binding motif (RNP1, RRM) protein 3 (222026_at), score: 0.66 RIN2Ras and Rab interactor 2 (209684_at), score: 0.7 RNASEH2Bribonuclease H2, subunit B (219056_at), score: 0.64 RNF187ring finger protein 187 (212155_at), score: 0.67 RNF19Aring finger protein 19A (220483_s_at), score: -0.73 RPL31ribosomal protein L31 (221593_s_at), score: 0.88 RPL35Aribosomal protein L35a (215208_x_at), score: -0.68 RUNDC3BRUN domain containing 3B (215321_at), score: -0.95 SCAMP1secretory carrier membrane protein 1 (206667_s_at), score: -0.72 SCML2sex comb on midleg-like 2 (Drosophila) (206147_x_at), score: -0.72 SENP3SUMO1/sentrin/SMT3 specific peptidase 3 (215113_s_at), score: -0.9 SH2D3ASH2 domain containing 3A (222169_x_at), score: -0.86 SLC11A1solute carrier family 11 (proton-coupled divalent metal ion transporters), member 1 (217473_x_at), score: -0.85 SLC25A16solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 (210686_x_at), score: -0.67 SLC25A20solute carrier family 25 (carnitine/acylcarnitine translocase), member 20 (203658_at), score: -0.66 SLC25A36solute carrier family 25, member 36 (201918_at), score: -0.63 SLC25A37solute carrier family 25, member 37 (218136_s_at), score: -0.9 SLC30A1solute carrier family 30 (zinc transporter), member 1 (212907_at), score: 0.66 SLC6A6solute carrier family 6 (neurotransmitter transporter, taurine), member 6 (205921_s_at), score: -0.69 SLC9A2solute carrier family 9 (sodium/hydrogen exchanger), member 2 (208039_at), score: -0.64 SMAD5SMAD family member 5 (205187_at), score: -0.68 SMEK1SMEK homolog 1, suppressor of mek1 (Dictyostelium) (220368_s_at), score: -0.94 SMG1SMG1 homolog, phosphatidylinositol 3-kinase-related kinase (C. elegans) (210057_at), score: -0.76 SMURF2SMAD specific E3 ubiquitin protein ligase 2 (205596_s_at), score: 0.83 SNRPD1small nuclear ribonucleoprotein D1 polypeptide 16kDa (202691_at), score: 0.72 SP110SP110 nuclear body protein (209761_s_at), score: -0.69 SPHARS-phase response (cyclin-related) (206272_at), score: 0.71 SRBD1S1 RNA binding domain 1 (219055_at), score: 0.69 SS18synovial sarcoma translocation, chromosome 18 (216684_s_at), score: -0.75 SSR3signal sequence receptor, gamma (translocon-associated protein gamma) (217790_s_at), score: 0.88 STARD7StAR-related lipid transfer (START) domain containing 7 (200028_s_at), score: 0.62 STAU2staufen, RNA binding protein, homolog 2 (Drosophila) (204226_at), score: 0.62 STK39serine threonine kinase 39 (STE20/SPS1 homolog, yeast) (202786_at), score: 0.73 STX16syntaxin 16 (221499_s_at), score: -0.72 SYPL1synaptophysin-like 1 (201259_s_at), score: -0.81 TAF1CTATA box binding protein (TBP)-associated factor, RNA polymerase I, C, 110kDa (203937_s_at), score: -0.69 TAF9BTAF9B RNA polymerase II, TATA box binding protein (TBP)-associated factor, 31kDa (221618_s_at), score: -0.7 TBC1D3TBC1 domain family, member 3 (209403_at), score: -0.68 TBX6T-box 6 (207684_at), score: -0.64 THAP4THAP domain containing 4 (220417_s_at), score: -0.65 TM9SF4transmembrane 9 superfamily protein member 4 (212194_s_at), score: -0.64 TMEM2transmembrane protein 2 (218113_at), score: 0.62 TNFRSF9tumor necrosis factor receptor superfamily, member 9 (207536_s_at), score: -0.69 TRIM25tripartite motif-containing 25 (206911_at), score: -0.73 TSPYL1TSPY-like 1 (221493_at), score: 0.63 TSPYL5TSPY-like 5 (213122_at), score: 0.63 TSTthiosulfate sulfurtransferase (rhodanese) (209605_at), score: 0.63 UAP1UDP-N-acteylglucosamine pyrophosphorylase 1 (209340_at), score: 0.72 UBAP2ubiquitin associated protein 2 (219192_at), score: 0.77 UBAP2Lubiquitin associated protein 2-like (201378_s_at), score: -0.7 UBE2Hubiquitin-conjugating enzyme E2H (UBC8 homolog, yeast) (221962_s_at), score: -0.66 UGDHUDP-glucose dehydrogenase (203343_at), score: 0.66 UGT1A6UDP glucuronosyltransferase 1 family, polypeptide A6 (206094_x_at), score: -0.7 ULK1unc-51-like kinase 1 (C. elegans) (209333_at), score: -0.7 USF2upstream transcription factor 2, c-fos interacting (214879_x_at), score: -0.68 UXS1UDP-glucuronate decarboxylase 1 (219675_s_at), score: 0.62 VGLL3vestigial like 3 (Drosophila) (220327_at), score: 0.81 VPS39vacuolar protein sorting 39 homolog (S. cerevisiae) (212156_at), score: -0.69 WACWW domain containing adaptor with coiled-coil (219679_s_at), score: -0.79 WNK1WNK lysine deficient protein kinase 1 (211992_at), score: -0.66 ZFRzinc finger RNA binding protein (33148_at), score: -0.64 ZNF117zinc finger protein 117 (207605_x_at), score: -0.64 ZNF224zinc finger protein 224 (216983_s_at), score: -0.83 ZNF318zinc finger protein 318 (203521_s_at), score: -0.64 ZNF675zinc finger protein 675 (217547_x_at), score: -0.71
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
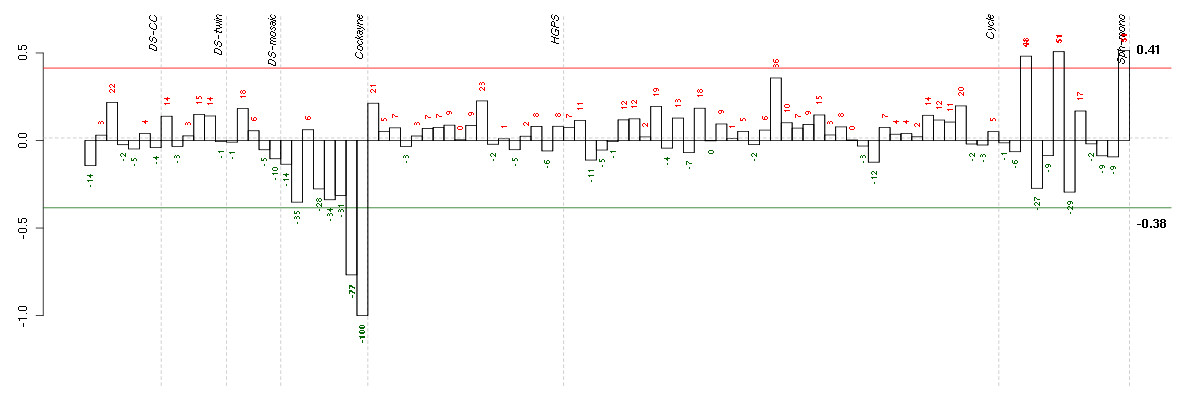
| E-GEOD-3407-raw-cel-1437949938.cel | 8 | 4 | Cockayne | hgu133a | none | CSB |
| E-GEOD-3407-raw-cel-1437949854.cel | 7 | 4 | Cockayne | hgu133a | CS | eGFP |
| E-GEOD-4219-raw-cel-1311956178.cel | 6 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956358.cel | 10 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956824.cel | 24 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |