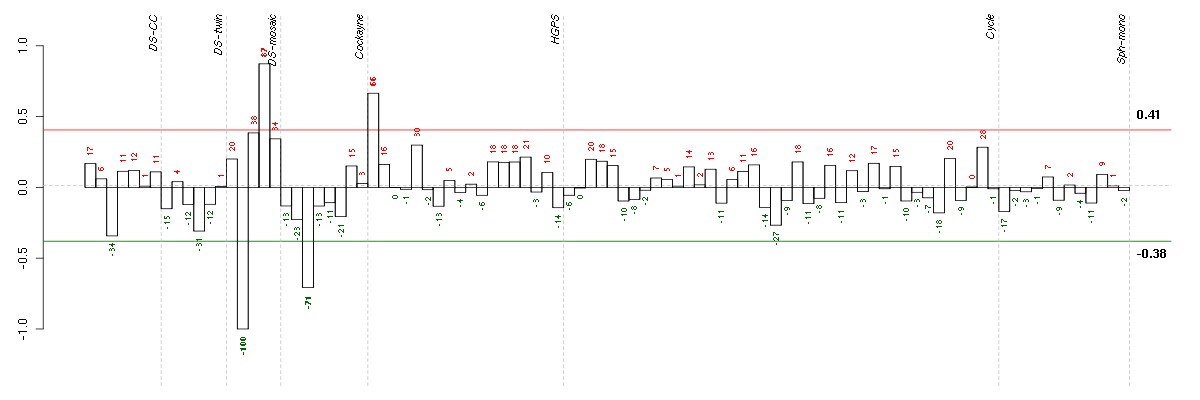

Under-expression is coded with green,
over-expression with red color.


plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
integral to membrane
Penetrating at least one phospholipid bilayer of a membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer. When used to describe a protein, indicates that all or part of the peptide sequence is embedded in the membrane.
extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
cytoplasm
All of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
Golgi apparatus
A compound membranous cytoplasmic organelle of eukaryotic cells, consisting of flattened, ribosome-free vesicles arranged in a more or less regular stack. The Golgi apparatus differs from the endoplasmic reticulum in often having slightly thicker membranes, appearing in sections as a characteristic shallow semicircle so that the convex side (cis or entry face) abuts the endoplasmic reticulum, secretory vesicles emerging from the concave side (trans or exit face). In vertebrate cells there is usually one such organelle, while in invertebrates and plants, where they are known usually as dictyosomes, there may be several scattered in the cytoplasm. The Golgi apparatus processes proteins produced on the ribosomes of the rough endoplasmic reticulum; such processing includes modification of the core oligosaccharides of glycoproteins, and the sorting and packaging of proteins for transport to a variety of cellular locations. Three different regions of the Golgi are now recognized both in terms of structure and function: cis, in the vicinity of the cis face, trans, in the vicinity of the trans face, and medial, lying between the cis and trans regions.
cytoplasmic membrane-bounded vesicle
A membrane-bounded vesicle found in the cytoplasm of the cell.
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
cytoplasmic vesicle
A vesicle formed of membrane or protein, found in the cytoplasm of a cell.
vesicle
Any small, fluid-filled, spherical organelle enclosed by membrane or protein.
membrane-bounded vesicle
Any small, fluid-filled, spherical organelle enclosed by a lipid bilayer.
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
cytoplasmic vesicle
A vesicle formed of membrane or protein, found in the cytoplasm of a cell.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
cytoplasmic membrane-bounded vesicle
A membrane-bounded vesicle found in the cytoplasm of the cell.
cytoplasmic membrane-bounded vesicle
A membrane-bounded vesicle found in the cytoplasm of the cell.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
Golgi apparatus
A compound membranous cytoplasmic organelle of eukaryotic cells, consisting of flattened, ribosome-free vesicles arranged in a more or less regular stack. The Golgi apparatus differs from the endoplasmic reticulum in often having slightly thicker membranes, appearing in sections as a characteristic shallow semicircle so that the convex side (cis or entry face) abuts the endoplasmic reticulum, secretory vesicles emerging from the concave side (trans or exit face). In vertebrate cells there is usually one such organelle, while in invertebrates and plants, where they are known usually as dictyosomes, there may be several scattered in the cytoplasm. The Golgi apparatus processes proteins produced on the ribosomes of the rough endoplasmic reticulum; such processing includes modification of the core oligosaccharides of glycoproteins, and the sorting and packaging of proteins for transport to a variety of cellular locations. Three different regions of the Golgi are now recognized both in terms of structure and function: cis, in the vicinity of the cis face, trans, in the vicinity of the trans face, and medial, lying between the cis and trans regions.
cytoplasmic vesicle
A vesicle formed of membrane or protein, found in the cytoplasm of a cell.

A4GALTalpha 1,4-galactosyltransferase (219488_at), score: 0.81 ADAM15ADAM metallopeptidase domain 15 (217007_s_at), score: 0.8 ADRBK1adrenergic, beta, receptor kinase 1 (201401_s_at), score: 0.71 AEBP1AE binding protein 1 (201792_at), score: 0.85 AESamino-terminal enhancer of split (217729_s_at), score: 0.56 AGPAT21-acylglycerol-3-phosphate O-acyltransferase 2 (lysophosphatidic acid acyltransferase, beta) (32837_at), score: 0.65 AGPAT31-acylglycerol-3-phosphate O-acyltransferase 3 (219723_x_at), score: 0.6 AGTPBP1ATP/GTP binding protein 1 (204500_s_at), score: -0.82 ANGPTL4angiopoietin-like 4 (221009_s_at), score: 0.71 ANTXR1anthrax toxin receptor 1 (220092_s_at), score: 0.57 AP1S1adaptor-related protein complex 1, sigma 1 subunit (205195_at), score: 0.79 APH1Aanterior pharynx defective 1 homolog A (C. elegans) (218389_s_at), score: 0.66 APOL6apolipoprotein L, 6 (219716_at), score: -0.89 APPL1adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 1 (218158_s_at), score: -0.79 ARFGAP1ADP-ribosylation factor GTPase activating protein 1 (217888_s_at), score: 0.7 ARFRP1ADP-ribosylation factor related protein 1 (215984_s_at), score: 0.66 ARHGDIARho GDP dissociation inhibitor (GDI) alpha (213606_s_at), score: 1 ARPC4actin related protein 2/3 complex, subunit 4, 20kDa (211672_s_at), score: 0.94 ASXL1additional sex combs like 1 (Drosophila) (212237_at), score: -0.76 ATP13A2ATPase type 13A2 (218608_at), score: 0.78 BACE2beta-site APP-cleaving enzyme 2 (217867_x_at), score: 0.57 BCL2L1BCL2-like 1 (215037_s_at), score: 0.77 BCL7AB-cell CLL/lymphoma 7A (203795_s_at), score: 0.57 BET1Lblocked early in transport 1 homolog (S. cerevisiae)-like (220470_at), score: 0.75 BRD2bromodomain containing 2 (214911_s_at), score: 0.67 BSGbasigin (Ok blood group) (208677_s_at), score: 0.72 BTBD2BTB (POZ) domain containing 2 (207722_s_at), score: 0.55 C19orf22chromosome 19 open reading frame 22 (221764_at), score: 0.87 C19orf6chromosome 19 open reading frame 6 (213986_s_at), score: 0.96 C19orf61chromosome 19 open reading frame 61 (221335_x_at), score: 0.58 C5orf23chromosome 5 open reading frame 23 (219054_at), score: 0.66 C9orf16chromosome 9 open reading frame 16 (204480_s_at), score: 0.72 CAMK2Bcalcium/calmodulin-dependent protein kinase II beta (34846_at), score: 0.59 CASP9caspase 9, apoptosis-related cysteine peptidase (203984_s_at), score: -0.77 CCND1cyclin D1 (208711_s_at), score: 0.76 CD44CD44 molecule (Indian blood group) (210916_s_at), score: 0.68 CDK10cyclin-dependent kinase 10 (210622_x_at), score: 0.56 CHKAcholine kinase alpha (204233_s_at), score: 0.68 CHST11carbohydrate (chondroitin 4) sulfotransferase 11 (219634_at), score: 0.68 CHST5carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 5 (219182_at), score: -0.83 CLPTM1cleft lip and palate associated transmembrane protein 1 (211136_s_at), score: 0.84 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: 0.69 COL1A1collagen, type I, alpha 1 (217430_x_at), score: 0.77 COL6A1collagen, type VI, alpha 1 (212940_at), score: 0.67 COL8A2collagen, type VIII, alpha 2 (221900_at), score: 0.6 COMPcartilage oligomeric matrix protein (205713_s_at), score: 0.66 COPAcoatomer protein complex, subunit alpha (214336_s_at), score: 0.59 CRIP2cysteine-rich protein 2 (208978_at), score: 0.58 CRLF1cytokine receptor-like factor 1 (206315_at), score: 0.71 CYBAcytochrome b-245, alpha polypeptide (203028_s_at), score: 0.59 CYR61cysteine-rich, angiogenic inducer, 61 (210764_s_at), score: 0.62 DAPK3death-associated protein kinase 3 (203890_s_at), score: 0.94 DCTN5dynactin 5 (p25) (209231_s_at), score: 0.63 DDA1DET1 and DDB1 associated 1 (218260_at), score: 0.66 DKK1dickkopf homolog 1 (Xenopus laevis) (204602_at), score: 0.65 DNM2dynamin 2 (202253_s_at), score: 0.68 DNMBPdynamin binding protein (212838_at), score: 0.64 DPM2dolichyl-phosphate mannosyltransferase polypeptide 2, regulatory subunit (209391_at), score: 0.55 DSG2desmoglein 2 (217901_at), score: 0.54 ECHDC2enoyl Coenzyme A hydratase domain containing 2 (218552_at), score: -0.85 EGFepidermal growth factor (beta-urogastrone) (206254_at), score: -0.95 EHD1EH-domain containing 1 (209037_s_at), score: 0.61 EHMT1euchromatic histone-lysine N-methyltransferase 1 (219339_s_at), score: 0.65 EIF4G1eukaryotic translation initiation factor 4 gamma, 1 (208624_s_at), score: 0.85 EIF5Aeukaryotic translation initiation factor 5A (201123_s_at), score: 0.83 ELK1ELK1, member of ETS oncogene family (210376_x_at), score: 0.82 ELP4elongation protein 4 homolog (S. cerevisiae) (203829_at), score: -0.78 EPHB4EPH receptor B4 (216680_s_at), score: 0.6 ESM1endothelial cell-specific molecule 1 (208394_x_at), score: 0.67 EZRezrin (208621_s_at), score: 0.83 FAM108A1family with sequence similarity 108, member A1 (221267_s_at), score: 0.57 FAM155Afamily with sequence similarity 155, member A (214825_at), score: 0.69 FASNfatty acid synthase (212218_s_at), score: 0.66 FBXL18F-box and leucine-rich repeat protein 18 (215068_s_at), score: 0.65 FGFR1fibroblast growth factor receptor 1 (210973_s_at), score: 0.86 FKBP8FK506 binding protein 8, 38kDa (208255_s_at), score: 0.57 FN1fibronectin 1 (214701_s_at), score: 0.6 FNBP4formin binding protein 4 (212232_at), score: -0.78 FRMD4AFERM domain containing 4A (208476_s_at), score: 0.74 FSCN1fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus) (210933_s_at), score: 0.81 FSTL3follistatin-like 3 (secreted glycoprotein) (203592_s_at), score: 0.58 FURINfurin (paired basic amino acid cleaving enzyme) (201945_at), score: 0.68 GAAglucosidase, alpha; acid (202812_at), score: 0.55 GALNT2UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-T2) (217787_s_at), score: 0.54 GBP2guanylate binding protein 2, interferon-inducible (202748_at), score: -0.77 GCAgrancalcin, EF-hand calcium binding protein (203765_at), score: -0.77 GLTPglycolipid transfer protein (219267_at), score: 0.71 GLULglutamate-ammonia ligase (glutamine synthetase) (200648_s_at), score: 0.56 GNAI2guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2 (201040_at), score: 0.62 GNB2guanine nucleotide binding protein (G protein), beta polypeptide 2 (200852_x_at), score: 0.58 GPR137G protein-coupled receptor 137 (43934_at), score: 0.58 GPR176G protein-coupled receptor 176 (206673_at), score: 0.56 GRAMD3GRAM domain containing 3 (218706_s_at), score: 0.54 HIST1H2BIhistone cluster 1, H2bi (208523_x_at), score: 0.71 HIST1H4Hhistone cluster 1, H4h (208180_s_at), score: 0.56 HIST2H2AA3histone cluster 2, H2aa3 (214290_s_at), score: 0.74 HMGA1high mobility group AT-hook 1 (210457_x_at), score: 0.75 HPS1Hermansky-Pudlak syndrome 1 (217354_s_at), score: 0.56 IFIT1interferon-induced protein with tetratricopeptide repeats 1 (203153_at), score: -0.78 IGFBP5insulin-like growth factor binding protein 5 (203424_s_at), score: 0.54 IL15interleukin 15 (205992_s_at), score: -0.83 IL15RAinterleukin 15 receptor, alpha (207375_s_at), score: -0.86 IL17RCinterleukin 17 receptor C (64440_at), score: 0.6 IL21Rinterleukin 21 receptor (219971_at), score: 0.57 IL4Rinterleukin 4 receptor (203233_at), score: 0.73 INHBAinhibin, beta A (210511_s_at), score: 0.6 IRF1interferon regulatory factor 1 (202531_at), score: -0.78 ISLRimmunoglobulin superfamily containing leucine-rich repeat (207191_s_at), score: 0.55 JAM3junctional adhesion molecule 3 (212813_at), score: 0.56 KCNN4potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 (204401_at), score: 0.62 KCTD20potassium channel tetramerisation domain containing 20 (214849_at), score: 0.58 KDELR1KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1 (200922_at), score: 0.82 KIAA0100KIAA0100 (201729_s_at), score: 0.77 KLF6Kruppel-like factor 6 (208961_s_at), score: 0.59 KLF7Kruppel-like factor 7 (ubiquitous) (204334_at), score: 0.71 LAMP1lysosomal-associated membrane protein 1 (201551_s_at), score: 0.83 LIMK2LIM domain kinase 2 (202193_at), score: 0.57 LMF1lipase maturation factor 1 (219135_s_at), score: 0.58 LOC100133105hypothetical protein LOC100133105 (214237_x_at), score: 0.8 LOXL2lysyl oxidase-like 2 (202997_s_at), score: 0.9 LSM12LSM12 homolog (S. cerevisiae) (212529_at), score: 0.64 LSSlanosterol synthase (2,3-oxidosqualene-lanosterol cyclase) (211019_s_at), score: 0.65 LTBP3latent transforming growth factor beta binding protein 3 (219922_s_at), score: 0.63 MAFKv-maf musculoaponeurotic fibrosarcoma oncogene homolog K (avian) (206750_at), score: 0.78 MAP1Bmicrotubule-associated protein 1B (214577_at), score: 0.55 MAP2K2mitogen-activated protein kinase kinase 2 (213490_s_at), score: 0.88 MAPKAPK2mitogen-activated protein kinase-activated protein kinase 2 (201461_s_at), score: 0.6 MAXMYC associated factor X (210734_x_at), score: 0.8 MAZMYC-associated zinc finger protein (purine-binding transcription factor) (207824_s_at), score: 0.65 MED31mediator complex subunit 31 (219318_x_at), score: 0.62 MFAP3Lmicrofibrillar-associated protein 3-like (205442_at), score: 0.63 MFN2mitofusin 2 (216205_s_at), score: 0.56 MGAT4Bmannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme B (220189_s_at), score: 0.67 MLXMAX-like protein X (217909_s_at), score: 0.65 MMP14matrix metallopeptidase 14 (membrane-inserted) (202827_s_at), score: 0.94 MPDU1mannose-P-dolichol utilization defect 1 (209208_at), score: 0.68 MUC1mucin 1, cell surface associated (207847_s_at), score: 0.64 MVDmevalonate (diphospho) decarboxylase (203027_s_at), score: 0.69 MYO10myosin X (201976_s_at), score: 0.65 NAB2NGFI-A binding protein 2 (EGR1 binding protein 2) (216017_s_at), score: 0.76 NAPAN-ethylmaleimide-sensitive factor attachment protein, alpha (206491_s_at), score: 0.6 NCLNnicalin homolog (zebrafish) (222206_s_at), score: 0.6 NCRNA00153non-protein coding RNA 153 (219961_s_at), score: -0.8 NFATC4nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 (205897_at), score: 0.72 NFICnuclear factor I/C (CCAAT-binding transcription factor) (206929_s_at), score: 0.61 NFKB2nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) (207535_s_at), score: 0.64 NRBP1nuclear receptor binding protein 1 (217765_at), score: 0.63 NTHL1nth endonuclease III-like 1 (E. coli) (209731_at), score: -0.82 NTMneurotrimin (222020_s_at), score: 0.86 NUCB1nucleobindin 1 (200646_s_at), score: 0.67 NUMA1nuclear mitotic apparatus protein 1 (214251_s_at), score: 0.68 OAZ2ornithine decarboxylase antizyme 2 (201364_s_at), score: 0.68 OPCMLopioid binding protein/cell adhesion molecule-like (214111_at), score: 0.82 ORAI2ORAI calcium release-activated calcium modulator 2 (218812_s_at), score: 0.56 P4HBprolyl 4-hydroxylase, beta polypeptide (200656_s_at), score: 0.65 PARP8poly (ADP-ribose) polymerase family, member 8 (219033_at), score: -0.9 PCDH9protocadherin 9 (219737_s_at), score: 0.65 PCDHGA11protocadherin gamma subfamily A, 11 (211876_x_at), score: 0.64 PCDHGA3protocadherin gamma subfamily A, 3 (216352_x_at), score: 0.74 PEX10peroxisomal biogenesis factor 10 (206351_s_at), score: 0.55 PFKLphosphofructokinase, liver (201102_s_at), score: 0.84 PIK3R2phosphoinositide-3-kinase, regulatory subunit 2 (beta) (207105_s_at), score: 0.54 PIONpigeon homolog (Drosophila) (222150_s_at), score: -0.95 PIP5K1Aphosphatidylinositol-4-phosphate 5-kinase, type I, alpha (211205_x_at), score: 0.71 PLD3phospholipase D family, member 3 (201050_at), score: 0.58 PLEC1plectin 1, intermediate filament binding protein 500kDa (216971_s_at), score: 1 POLDIP3polymerase (DNA-directed), delta interacting protein 3 (215357_s_at), score: 0.81 POLR2Epolymerase (RNA) II (DNA directed) polypeptide E, 25kDa (213887_s_at), score: 0.66 POLR2J2polymerase (RNA) II (DNA directed) polypeptide J2 (216242_x_at), score: 0.69 PPP2R1Aprotein phosphatase 2 (formerly 2A), regulatory subunit A, alpha isoform (200695_at), score: 0.77 PPP5Cprotein phosphatase 5, catalytic subunit (201979_s_at), score: 0.72 PRPF6PRP6 pre-mRNA processing factor 6 homolog (S. cerevisiae) (208879_x_at), score: 0.86 PRRX1paired related homeobox 1 (205991_s_at), score: 0.54 PSENENpresenilin enhancer 2 homolog (C. elegans) (218302_at), score: 0.56 PTBP1polypyrimidine tract binding protein 1 (212016_s_at), score: 0.81 PTGS2prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) (204748_at), score: 0.6 PTHLHparathyroid hormone-like hormone (211756_at), score: 0.55 PTMSparathymosin (218045_x_at), score: 0.87 PTP4A1protein tyrosine phosphatase type IVA, member 1 (200730_s_at), score: 0.59 PTPN14protein tyrosine phosphatase, non-receptor type 14 (205503_at), score: 0.62 PTRFpolymerase I and transcript release factor (208790_s_at), score: 0.77 PURApurine-rich element binding protein A (204021_s_at), score: 0.59 PXNpaxillin (211823_s_at), score: 0.79 RAB11BRAB11B, member RAS oncogene family (34478_at), score: 0.7 RAB35RAB35, member RAS oncogene family (205461_at), score: 0.77 RAB3BRAB3B, member RAS oncogene family (205924_at), score: 0.67 RANGAP1Ran GTPase activating protein 1 (212125_at), score: 0.71 RASA3RAS p21 protein activator 3 (206220_s_at), score: 0.57 RASA4RAS p21 protein activator 4 (208534_s_at), score: 0.96 RELAv-rel reticuloendotheliosis viral oncogene homolog A (avian) (209878_s_at), score: 0.84 RNASEH1ribonuclease H1 (218496_at), score: 0.55 RNF126ring finger protein 126 (205748_s_at), score: 0.8 RPS20ribosomal protein S20 (216247_at), score: 0.69 RUNDC3BRUN domain containing 3B (215321_at), score: -0.79 RXRBretinoid X receptor, beta (215099_s_at), score: 0.68 SAMD14sterile alpha motif domain containing 14 (213866_at), score: 0.6 SBF1SET binding factor 1 (39835_at), score: 0.56 SCAMP2secretory carrier membrane protein 2 (218143_s_at), score: 0.62 SDC4syndecan 4 (202071_at), score: 0.55 SEC14L2SEC14-like 2 (S. cerevisiae) (204541_at), score: 0.75 SEMA5Asema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A (205405_at), score: 0.8 SEPW1selenoprotein W, 1 (201194_at), score: 0.56 SERPINE2serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 (212190_at), score: 0.56 SETD8SET domain containing (lysine methyltransferase) 8 (220200_s_at), score: 0.57 SH3GLB2SH3-domain GRB2-like endophilin B2 (218813_s_at), score: 0.71 SH3PXD2ASH3 and PX domains 2A (213252_at), score: 0.81 SHC1SHC (Src homology 2 domain containing) transforming protein 1 (201469_s_at), score: 0.69 SIRT1sirtuin (silent mating type information regulation 2 homolog) 1 (S. cerevisiae) (218878_s_at), score: -0.83 SLC12A4solute carrier family 12 (potassium/chloride transporters), member 4 (209401_s_at), score: 0.67 SLC16A3solute carrier family 16, member 3 (monocarboxylic acid transporter 4) (202855_s_at), score: 0.82 SLC17A5solute carrier family 17 (anion/sugar transporter), member 5 (221041_s_at), score: 0.6 SLC35C1solute carrier family 35, member C1 (218485_s_at), score: 0.65 SLC35E1solute carrier family 35, member E1 (222263_at), score: 0.82 SLC46A3solute carrier family 46, member 3 (214719_at), score: 0.55 SLC5A3solute carrier family 5 (sodium/myo-inositol cotransporter), member 3 (213164_at), score: 0.71 SLC6A9solute carrier family 6 (neurotransmitter transporter, glycine), member 9 (207043_s_at), score: 0.71 SLFN12schlafen family member 12 (219885_at), score: -0.79 SMARCA4SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 (212520_s_at), score: 0.6 SNNstannin (218032_at), score: -0.77 SPPL2Bsignal peptide peptidase-like 2B (215833_s_at), score: 0.72 SRPRsignal recognition particle receptor (docking protein) (200917_s_at), score: 0.84 SSBP2single-stranded DNA binding protein 2 (203787_at), score: -0.76 SSR3signal sequence receptor, gamma (translocon-associated protein gamma) (217790_s_at), score: 0.58 ST6GALNAC4ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 (221551_x_at), score: 0.74 STARD3StAR-related lipid transfer (START) domain containing 3 (202991_at), score: 0.59 STIP1stress-induced-phosphoprotein 1 (212009_s_at), score: 0.57 SULT1A2sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2 (207122_x_at), score: -0.98 SYT1synaptotagmin I (203999_at), score: -0.84 TAF4TAF4 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 135kDa (213090_s_at), score: -0.97 TBC1D12TBC1 domain family, member 12 (221858_at), score: -0.77 TGFB1transforming growth factor, beta 1 (203085_s_at), score: 0.56 THY1Thy-1 cell surface antigen (208850_s_at), score: 0.55 TLR3toll-like receptor 3 (206271_at), score: -0.87 TM9SF4transmembrane 9 superfamily protein member 4 (212194_s_at), score: 0.57 TMEM204transmembrane protein 204 (219315_s_at), score: 0.55 TMEM97transmembrane protein 97 (212279_at), score: -0.8 TNFRSF10Dtumor necrosis factor receptor superfamily, member 10d, decoy with truncated death domain (210654_at), score: 0.63 TNFRSF12Atumor necrosis factor receptor superfamily, member 12A (218368_s_at), score: 0.69 TNFRSF1Btumor necrosis factor receptor superfamily, member 1B (203508_at), score: -0.78 TOP3Atopoisomerase (DNA) III alpha (214300_s_at), score: 0.7 TRAM2translocation associated membrane protein 2 (202368_s_at), score: 0.66 TRIM25tripartite motif-containing 25 (206911_at), score: 0.58 TRMT2BTRM2 tRNA methyltransferase 2 homolog B (S. cerevisiae) (205238_at), score: 0.57 TSPAN4tetraspanin 4 (209264_s_at), score: 0.57 UBE2Mubiquitin-conjugating enzyme E2M (UBC12 homolog, yeast) (203109_at), score: 0.78 UBXN6UBX domain protein 6 (220757_s_at), score: 0.56 WDR1WD repeat domain 1 (210935_s_at), score: 0.63 XYLT1xylosyltransferase I (213725_x_at), score: 0.69 YIPF5Yip1 domain family, member 5 (221423_s_at), score: 0.63 YWHAEtyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide (210317_s_at), score: 0.54 ZER1zer-1 homolog (C. elegans) (202456_s_at), score: 0.64 ZFPL1zinc finger protein-like 1 (209428_s_at), score: 0.84 ZNF185zinc finger protein 185 (LIM domain) (203585_at), score: 0.61 ZNF217zinc finger protein 217 (203739_at), score: 0.69 ZRANB1zinc finger, RAN-binding domain containing 1 (201219_at), score: -0.84
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| 46B.CEL | 2 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 2 |
| E-GEOD-3407-raw-cel-1437949655.cel | 3 | 4 | Cockayne | hgu133a | none | CSB |
| E-GEOD-3860-raw-cel-1561690199.cel | 1 | 5 | HGPS | hgu133a | none | GM0316B |
| 47B.CEL | 4 | 3 | DS-mosaic | hgu133plus2 | Down mosaic | DS-mosaic 4 |