
Under-expression is coded with green,
over-expression with red color.

multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
tissue development
The process whose specific outcome is the progression of a tissue over time, from its formation to the mature structure.
ectoderm development
The process whose specific outcome is the progression of the ectoderm over time, from its formation to the mature structure. In animal embryos, the ectoderm is the outer germ layer of the embryo, formed during gastrulation.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
epidermis development
The process whose specific outcome is the progression of the epidermis over time, from its formation to the mature structure. The epidermis is the outer epithelial layer of a plant or animal, it may be a single layer that produces an extracellular material (e.g. the cuticle of arthropods) or a complex stratified squamous epithelium, as in the case of many vertebrate species.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
developmental process
A biological process whose specific outcome is the progression of an integrated living unit: an anatomical structure (which may be a subcellular structure, cell, tissue, or organ), or organism over time from an initial condition to a later condition.
organ development
Development of a tissue or tissues that work together to perform a specific function or functions. Development pertains to the process whose specific outcome is the progression of a structure over time, from its formation to the mature structure. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
anatomical structure development
The biological process whose specific outcome is the progression of an anatomical structure from an initial condition to its mature state. This process begins with the formation of the structure and ends with the mature structure, whatever form that may be including its natural destruction. An anatomical structure is any biological entity that occupies space and is distinguished from its surroundings. Anatomical structures can be macroscopic such as a carpel, or microscopic such as an acrosome.
all
This term is the most general term possible
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
organ development
Development of a tissue or tissues that work together to perform a specific function or functions. Development pertains to the process whose specific outcome is the progression of a structure over time, from its formation to the mature structure. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
tissue development
The process whose specific outcome is the progression of a tissue over time, from its formation to the mature structure.
epidermis development
The process whose specific outcome is the progression of the epidermis over time, from its formation to the mature structure. The epidermis is the outer epithelial layer of a plant or animal, it may be a single layer that produces an extracellular material (e.g. the cuticle of arthropods) or a complex stratified squamous epithelium, as in the case of many vertebrate species.

extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
extracellular space
That part of a multicellular organism outside the cells proper, usually taken to be outside the plasma membranes, and occupied by fluid.
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
all
This term is the most general term possible
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.

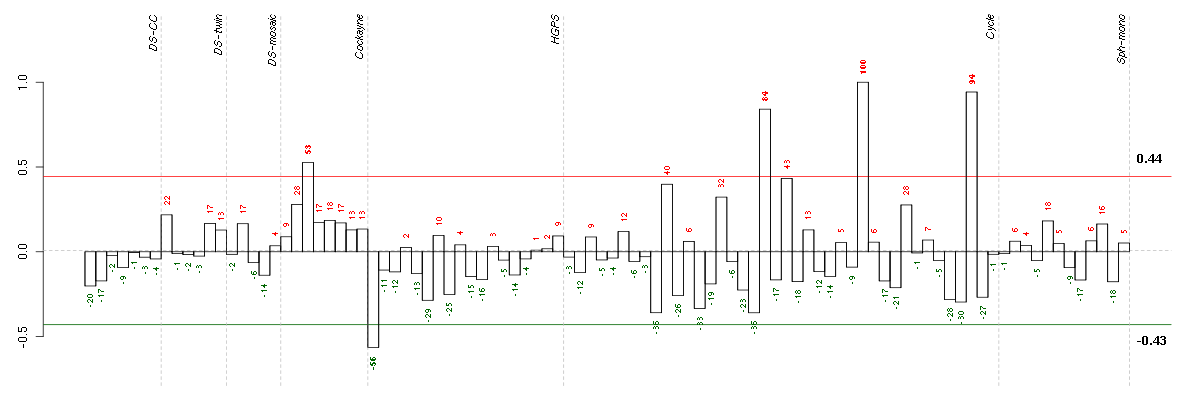
ABCA2ATP-binding cassette, sub-family A (ABC1), member 2 (212772_s_at), score: -0.46 ADAMTS6ADAM metallopeptidase with thrombospondin type 1 motif, 6 (220866_at), score: -0.49 AFG3L2AFG3 ATPase family gene 3-like 2 (yeast) (202486_at), score: 0.61 AGBL5ATP/GTP binding protein-like 5 (218480_at), score: 0.71 ALDH3A2aldehyde dehydrogenase 3 family, member A2 (202053_s_at), score: 0.62 ANKRD28ankyrin repeat domain 28 (213035_at), score: 0.68 ANXA10annexin A10 (210143_at), score: -0.56 AOF2amine oxidase (flavin containing) domain 2 (212348_s_at), score: 0.68 APOBEC3Fapolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3F (214995_s_at), score: 0.65 ARFGAP1ADP-ribosylation factor GTPase activating protein 1 (217888_s_at), score: -0.44 ARL17ADP-ribosylation factor-like 17 (210435_at), score: -0.53 ARNT2aryl-hydrocarbon receptor nuclear translocator 2 (202986_at), score: 0.66 ARNTLaryl hydrocarbon receptor nuclear translocator-like (209824_s_at), score: -0.49 ASB1ankyrin repeat and SOCS box-containing 1 (212819_at), score: -0.52 ASPNasporin (219087_at), score: -0.53 ATP13A2ATPase type 13A2 (218608_at), score: -0.45 B3GNTL1UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase-like 1 (213589_s_at), score: 0.92 BAALCbrain and acute leukemia, cytoplasmic (218899_s_at), score: -0.51 BCL10B-cell CLL/lymphoma 10 (205263_at), score: -0.43 BCS1LBCS1-like (yeast) (207618_s_at), score: 0.67 BMP6bone morphogenetic protein 6 (206176_at), score: -0.5 BTN3A1butyrophilin, subfamily 3, member A1 (209770_at), score: 0.59 C11orf67chromosome 11 open reading frame 67 (221600_s_at), score: 0.65 C12orf52chromosome 12 open reading frame 52 (221777_at), score: 0.66 C14orf45chromosome 14 open reading frame 45 (220173_at), score: -0.52 C17orf39chromosome 17 open reading frame 39 (220058_at), score: 0.79 C17orf90chromosome 17 open reading frame 90 (50374_at), score: 0.62 C19orf22chromosome 19 open reading frame 22 (221764_at), score: -0.6 C2CD2LC2CD2-like (204757_s_at), score: -0.7 C2orf49chromosome 2 open reading frame 49 (219662_at), score: 0.6 C5orf23chromosome 5 open reading frame 23 (219054_at), score: -0.6 C6orf62chromosome 6 open reading frame 62 (213875_x_at), score: 0.75 C9orf114chromosome 9 open reading frame 114 (218565_at), score: 0.9 CALB2calbindin 2 (205428_s_at), score: -0.52 CAMK2Bcalcium/calmodulin-dependent protein kinase II beta (34846_at), score: -0.43 CCL11chemokine (C-C motif) ligand 11 (210133_at), score: -0.46 CD320CD320 molecule (218529_at), score: 0.62 CD55CD55 molecule, decay accelerating factor for complement (Cromer blood group) (201925_s_at), score: -0.55 CDC2L6cell division cycle 2-like 6 (CDK8-like) (212899_at), score: 0.61 CDCP1CUB domain containing protein 1 (218451_at), score: -0.59 CDK5cyclin-dependent kinase 5 (204247_s_at), score: 0.6 CENPTcentromere protein T (218148_at), score: -0.46 CEP70centrosomal protein 70kDa (219036_at), score: 0.67 CHPFchondroitin polymerizing factor (202175_at), score: -0.43 CLCA2chloride channel accessory 2 (206165_s_at), score: -0.57 CLCF1cardiotrophin-like cytokine factor 1 (219500_at), score: -0.51 CLEC3BC-type lectin domain family 3, member B (205200_at), score: -0.79 CLK1CDC-like kinase 1 (214683_s_at), score: -0.56 CLUclusterin (208791_at), score: -0.65 CNNM2cyclin M2 (206818_s_at), score: 0.67 CNPY3canopy 3 homolog (zebrafish) (217931_at), score: 0.59 COL5A3collagen, type V, alpha 3 (52255_s_at), score: -0.77 COL7A1collagen, type VII, alpha 1 (217312_s_at), score: -0.51 COL8A2collagen, type VIII, alpha 2 (221900_at), score: -0.52 COMPcartilage oligomeric matrix protein (205713_s_at), score: -0.53 COQ3coenzyme Q3 homolog, methyltransferase (S. cerevisiae) (221227_x_at), score: 0.61 CORO2Bcoronin, actin binding protein, 2B (209789_at), score: -0.74 CRABP2cellular retinoic acid binding protein 2 (202575_at), score: -0.46 CST6cystatin E/M (206595_at), score: -0.69 CTNNBIP1catenin, beta interacting protein 1 (203081_at), score: -0.44 CTSOcathepsin O (203758_at), score: -0.5 CXCL6chemokine (C-X-C motif) ligand 6 (granulocyte chemotactic protein 2) (206336_at), score: -0.59 DARS2aspartyl-tRNA synthetase 2, mitochondrial (218365_s_at), score: 0.62 DCLK1doublecortin-like kinase 1 (205399_at), score: -0.43 DENND5BDENN/MADD domain containing 5B (215058_at), score: -0.53 DIRAS3DIRAS family, GTP-binding RAS-like 3 (215506_s_at), score: -0.75 DNAJC8DnaJ (Hsp40) homolog, subfamily C, member 8 (205545_x_at), score: 0.62 DUS4Ldihydrouridine synthase 4-like (S. cerevisiae) (205761_s_at), score: 0.72 DUSP5dual specificity phosphatase 5 (209457_at), score: -0.64 DUSP6dual specificity phosphatase 6 (208891_at), score: -0.56 DZIP3DAZ interacting protein 3, zinc finger (213186_at), score: 0.63 EIF3Heukaryotic translation initiation factor 3, subunit H (201592_at), score: 0.65 ELNelastin (212670_at), score: -0.45 EMCNendomucin (219436_s_at), score: 0.62 EML1echinoderm microtubule associated protein like 1 (204797_s_at), score: -0.49 EMP1epithelial membrane protein 1 (201325_s_at), score: -0.5 ERO1LERO1-like (S. cerevisiae) (218498_s_at), score: -0.54 ESM1endothelial cell-specific molecule 1 (208394_x_at), score: -0.61 ESRRAestrogen-related receptor alpha (1487_at), score: -0.62 EVI2Aecotropic viral integration site 2A (204774_at), score: 0.81 EVI2Becotropic viral integration site 2B (211742_s_at), score: 0.78 EXOSC5exosome component 5 (218481_at), score: 0.76 FAM174Bfamily with sequence similarity 174, member B (51158_at), score: -0.43 FAM48Afamily with sequence similarity 48, member A (221774_x_at), score: 0.65 FAM63Bfamily with sequence similarity 63, member B (214691_x_at), score: -0.76 FBXO31F-box protein 31 (219785_s_at), score: 0.82 FEM1Bfem-1 homolog b (C. elegans) (212367_at), score: -0.46 FEM1Cfem-1 homolog c (C. elegans) (213341_at), score: -0.56 FERMT1fermitin family homolog 1 (Drosophila) (218796_at), score: -0.44 FGFR2fibroblast growth factor receptor 2 (208229_at), score: 0.61 FKSG49FKSG49 (211454_x_at), score: 0.6 GAAglucosidase, alpha; acid (202812_at), score: -0.67 GABBR2gamma-aminobutyric acid (GABA) B receptor, 2 (209990_s_at), score: -0.5 GANgigaxonin (220124_at), score: -0.71 GARNL1GTPase activating Rap/RanGAP domain-like 1 (214855_s_at), score: -0.47 GAS7growth arrest-specific 7 (202191_s_at), score: -0.55 GIT1G protein-coupled receptor kinase interacting ArfGAP 1 (218030_at), score: -0.49 GNB1Lguanine nucleotide binding protein (G protein), beta polypeptide 1-like (220762_s_at), score: 0.69 GNPDA1glucosamine-6-phosphate deaminase 1 (202382_s_at), score: 0.61 GOLPH3golgi phosphoprotein 3 (coat-protein) (217803_at), score: -0.55 GPM6Bglycoprotein M6B (209167_at), score: -0.52 GPR153G protein-coupled receptor 153 (221902_at), score: -0.64 GPR177G protein-coupled receptor 177 (221958_s_at), score: -0.5 GPR27G protein-coupled receptor 27 (221306_at), score: 0.62 GPRASP1G protein-coupled receptor associated sorting protein 1 (204793_at), score: 0.79 GRB14growth factor receptor-bound protein 14 (206204_at), score: -0.54 HERC3hect domain and RLD 3 (206183_s_at), score: -0.46 HGFhepatocyte growth factor (hepapoietin A; scatter factor) (209960_at), score: -0.56 HINFPhistone H4 transcription factor (206495_s_at), score: 0.67 HIPK3homeodomain interacting protein kinase 3 (210148_at), score: -0.45 HIST1H2AGhistone cluster 1, H2ag (207156_at), score: 0.68 HIST1H2BIhistone cluster 1, H2bi (208523_x_at), score: -0.48 HIST1H4Hhistone cluster 1, H4h (208180_s_at), score: -0.54 HTR2A5-hydroxytryptamine (serotonin) receptor 2A (207135_at), score: -0.55 IER2immediate early response 2 (202081_at), score: -0.55 IGF2insulin-like growth factor 2 (somatomedin A) (202409_at), score: -0.49 IL13RA2interleukin 13 receptor, alpha 2 (206172_at), score: -0.66 IL16interleukin 16 (lymphocyte chemoattractant factor) (209827_s_at), score: -0.43 IL33interleukin 33 (209821_at), score: -0.77 INSIG2insulin induced gene 2 (209566_at), score: -0.5 INTS8integrator complex subunit 8 (218905_at), score: 0.69 KCNE4potassium voltage-gated channel, Isk-related family, member 4 (222379_at), score: 0.59 KCTD12potassium channel tetramerisation domain containing 12 (212192_at), score: -0.48 KIAA0141KIAA0141 (201977_s_at), score: 0.6 KIAA0247KIAA0247 (202181_at), score: -0.51 KIAA0586KIAA0586 (205631_at), score: 0.74 KLHL5kelch-like 5 (Drosophila) (220682_s_at), score: 0.69 KRT14keratin 14 (209351_at), score: -0.58 KRT19keratin 19 (201650_at), score: -0.56 KRT33Akeratin 33A (208483_x_at), score: -0.63 LANCL2LanC lantibiotic synthetase component C-like 2 (bacterial) (218219_s_at), score: 0.62 LARGElike-glycosyltransferase (215543_s_at), score: -0.43 LMAN2Llectin, mannose-binding 2-like (221274_s_at), score: 0.68 LPCAT3lysophosphatidylcholine acyltransferase 3 (202793_at), score: -0.56 LRP1low density lipoprotein-related protein 1 (alpha-2-macroglobulin receptor) (200785_s_at), score: -0.5 LRP2low density lipoprotein-related protein 2 (205710_at), score: -0.43 LTBP3latent transforming growth factor beta binding protein 3 (219922_s_at), score: -0.46 MAN1B1mannosidase, alpha, class 1B, member 1 (65884_at), score: -0.45 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: -0.48 MAP7D3MAP7 domain containing 3 (219576_at), score: 0.67 MAST2microtubule associated serine/threonine kinase 2 (211593_s_at), score: -0.46 MBmyoglobin (204179_at), score: -0.43 MED31mediator complex subunit 31 (219318_x_at), score: -0.45 MEGF6multiple EGF-like-domains 6 (213942_at), score: -0.55 MMP10matrix metallopeptidase 10 (stromelysin 2) (205680_at), score: -0.54 MMP16matrix metallopeptidase 16 (membrane-inserted) (207012_at), score: -0.6 MMP24matrix metallopeptidase 24 (membrane-inserted) (49679_s_at), score: -0.43 MRPL34mitochondrial ribosomal protein L34 (221692_s_at), score: 0.63 MRPL52mitochondrial ribosomal protein L52 (221997_s_at), score: 0.64 MSNmoesin (200600_at), score: -0.51 MSTP9macrophage stimulating, pseudogene 9 (213382_at), score: -0.49 MTAPmethylthioadenosine phosphorylase (211363_s_at), score: -0.45 MUC1mucin 1, cell surface associated (207847_s_at), score: -0.45 MXRA5matrix-remodelling associated 5 (209596_at), score: -0.5 N6AMT1N-6 adenine-specific DNA methyltransferase 1 (putative) (220311_at), score: 0.6 NAGPAN-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase (205090_s_at), score: 0.69 NARS2asparaginyl-tRNA synthetase 2, mitochondrial (putative) (219217_at), score: 0.67 NBL1neuroblastoma, suppression of tumorigenicity 1 (37005_at), score: -0.46 NCALDneurocalcin delta (211685_s_at), score: -0.47 NDUFA2NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2, 8kDa (213550_s_at), score: 0.76 NEFLneurofilament, light polypeptide (221805_at), score: -0.53 NFATC2IPnuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 interacting protein (217526_at), score: -0.45 NFATC4nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 (205897_at), score: -0.43 NMBneuromedin B (205204_at), score: 0.82 NOL8nucleolar protein 8 (218244_at), score: 0.7 NPR3natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) (219789_at), score: -0.56 NSUN5BNOL1/NOP2/Sun domain family, member 5B (214100_x_at), score: 0.63 NTHL1nth endonuclease III-like 1 (E. coli) (209731_at), score: 0.76 NTMneurotrimin (222020_s_at), score: -0.51 NUCKS1nuclear casein kinase and cyclin-dependent kinase substrate 1 (222027_at), score: 0.65 NUDT1nudix (nucleoside diphosphate linked moiety X)-type motif 1 (204766_s_at), score: 0.63 NUDT6nudix (nucleoside diphosphate linked moiety X)-type motif 6 (220183_s_at), score: 0.82 OLFM1olfactomedin 1 (213131_at), score: -0.49 OLFML2Aolfactomedin-like 2A (213075_at), score: -0.47 OPCMLopioid binding protein/cell adhesion molecule-like (214111_at), score: -0.69 P2RX5purinergic receptor P2X, ligand-gated ion channel, 5 (210448_s_at), score: -0.52 PAFAH1B3platelet-activating factor acetylhydrolase, isoform Ib, gamma subunit 29kDa (203228_at), score: 0.68 PAN2PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) (203117_s_at), score: 0.62 PCDH9protocadherin 9 (219737_s_at), score: -0.51 PDLIM4PDZ and LIM domain 4 (211564_s_at), score: -0.45 PENKproenkephalin (213791_at), score: -0.63 PID1phosphotyrosine interaction domain containing 1 (219093_at), score: -0.43 PIGLphosphatidylinositol glycan anchor biosynthesis, class L (213889_at), score: 0.68 PIP5K1Cphosphatidylinositol-4-phosphate 5-kinase, type I, gamma (212518_at), score: -0.44 PKIAprotein kinase (cAMP-dependent, catalytic) inhibitor alpha (204612_at), score: -0.51 PLAURplasminogen activator, urokinase receptor (211924_s_at), score: -0.63 PLCXD1phosphatidylinositol-specific phospholipase C, X domain containing 1 (218951_s_at), score: -0.52 PLXNB1plexin B1 (215807_s_at), score: -0.58 POU4F1POU class 4 homeobox 1 (206940_s_at), score: -0.48 PPFIBP1PTPRF interacting protein, binding protein 1 (liprin beta 1) (214374_s_at), score: 0.6 PRELPproline/arginine-rich end leucine-rich repeat protein (204223_at), score: -0.58 PRKG2protein kinase, cGMP-dependent, type II (207505_at), score: -0.55 PRSS2protease, serine, 2 (trypsin 2) (205402_x_at), score: -0.48 PRSS3protease, serine, 3 (213421_x_at), score: -0.73 PSD4pleckstrin and Sec7 domain containing 4 (203317_at), score: -0.62 PSMB9proteasome (prosome, macropain) subunit, beta type, 9 (large multifunctional peptidase 2) (204279_at), score: 0.6 PTCD1pentatricopeptide repeat domain 1 (218956_s_at), score: 0.69 PTENP1phosphatase and tensin homolog pseudogene 1 (217492_s_at), score: 0.66 PTGS1prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) (215813_s_at), score: -0.68 PTGS2prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) (204748_at), score: -0.63 PTHLHparathyroid hormone-like hormone (211756_at), score: -0.46 PTPRBprotein tyrosine phosphatase, receptor type, B (205846_at), score: -0.65 PTTG3pituitary tumor-transforming 3 (208511_at), score: 0.63 PXMP4peroxisomal membrane protein 4, 24kDa (219428_s_at), score: 0.63 PYCARDPYD and CARD domain containing (221666_s_at), score: 0.62 QSOX1quiescin Q6 sulfhydryl oxidase 1 (201482_at), score: -0.5 RAB6CRAB6C, member RAS oncogene family (210406_s_at), score: -0.56 RAB8BRAB8B, member RAS oncogene family (219210_s_at), score: 0.62 RAPGEF6Rap guanine nucleotide exchange factor (GEF) 6 (219112_at), score: -0.49 RASA2RAS p21 protein activator 2 (206636_at), score: -0.44 RCC1regulator of chromosome condensation 1 (206499_s_at), score: 0.62 RFX5regulatory factor X, 5 (influences HLA class II expression) (202963_at), score: 0.62 RFX7regulatory factor X, 7 (218430_s_at), score: 0.69 RHBDD3rhomboid domain containing 3 (204402_at), score: -0.49 RHBDF2rhomboid 5 homolog 2 (Drosophila) (219202_at), score: -0.48 RIPK1receptor (TNFRSF)-interacting serine-threonine kinase 1 (209941_at), score: -0.52 RNF126ring finger protein 126 (205748_s_at), score: -0.5 RPL14ribosomal protein L14 (219138_at), score: 0.83 RPL27Aribosomal protein L27a (212044_s_at), score: 0.96 RPL38ribosomal protein L38 (202028_s_at), score: 0.77 RPLP2ribosomal protein, large, P2 (200908_s_at), score: 0.68 RPP40ribonuclease P/MRP 40kDa subunit (213427_at), score: 0.61 RPS11ribosomal protein S11 (213350_at), score: 0.96 RPS19ribosomal protein S19 (202648_at), score: 0.73 SAT1spermidine/spermine N1-acetyltransferase 1 (213988_s_at), score: -0.49 SDR39U1short chain dehydrogenase/reductase family 39U, member 1 (213398_s_at), score: 0.69 SEC14L2SEC14-like 2 (S. cerevisiae) (204541_at), score: -0.5 SERPINB9serpin peptidase inhibitor, clade B (ovalbumin), member 9 (209723_at), score: 0.6 SERPINE2serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 (212190_at), score: -0.54 SERPING1serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 (200986_at), score: -0.45 SETMARSET domain and mariner transposase fusion gene (206554_x_at), score: 0.85 SGCDsarcoglycan, delta (35kDa dystrophin-associated glycoprotein) (213543_at), score: -0.48 SGK3serum/glucocorticoid regulated kinase family, member 3 (220038_at), score: -0.43 SIPA1L3signal-induced proliferation-associated 1 like 3 (213600_at), score: -0.52 SLC20A2solute carrier family 20 (phosphate transporter), member 2 (202744_at), score: -0.53 SLC38A4solute carrier family 38, member 4 (220786_s_at), score: 0.72 SLIT3slit homolog 3 (Drosophila) (203813_s_at), score: -0.44 SMU1smu-1 suppressor of mec-8 and unc-52 homolog (C. elegans) (218393_s_at), score: 0.64 SNRPEsmall nuclear ribonucleoprotein polypeptide E (215450_at), score: 0.7 SNX13sorting nexin 13 (213292_s_at), score: 0.67 SOX9SRY (sex determining region Y)-box 9 (202935_s_at), score: -0.5 SPA17sperm autoantigenic protein 17 (205406_s_at), score: 0.73 SPAG8sperm associated antigen 8 (206816_s_at), score: 0.64 SPP1secreted phosphoprotein 1 (209875_s_at), score: 0.65 SPRY4sprouty homolog 4 (Drosophila) (221489_s_at), score: -0.5 STC1stanniocalcin 1 (204597_x_at), score: -0.58 STX3syntaxin 3 (209238_at), score: 0.61 SULT1A2sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2 (207122_x_at), score: 0.6 SYNMsynemin, intermediate filament protein (212730_at), score: 0.62 SYT11synaptotagmin XI (209198_s_at), score: 0.74 TAPBPTAP binding protein (tapasin) (208829_at), score: -0.43 TFDP2transcription factor Dp-2 (E2F dimerization partner 2) (203588_s_at), score: 0.69 TGFAtransforming growth factor, alpha (205016_at), score: -0.59 TGFBRAP1transforming growth factor, beta receptor associated protein 1 (205210_at), score: 0.69 THOC2THO complex 2 (212994_at), score: 0.7 THYN1thymocyte nuclear protein 1 (218491_s_at), score: 0.68 TIMP3TIMP metallopeptidase inhibitor 3 (201149_s_at), score: -0.44 TMEM106Ctransmembrane protein 106C (201764_at), score: 0.64 TMEM158transmembrane protein 158 (213338_at), score: -0.52 TMSB15Athymosin beta 15a (205347_s_at), score: 0.81 TNFAIP3tumor necrosis factor, alpha-induced protein 3 (202644_s_at), score: -0.49 TOXthymocyte selection-associated high mobility group box (204529_s_at), score: 0.61 TP53TG1TP53 target 1 (non-protein coding) (209917_s_at), score: 0.73 TPK1thiamin pyrophosphokinase 1 (221218_s_at), score: -0.77 TPMTthiopurine S-methyltransferase (203672_x_at), score: 0.63 TRMT61BtRNA methyltransferase 61 homolog B (S. cerevisiae) (221229_s_at), score: 0.68 TRY6trypsinogen C (215395_x_at), score: -0.5 TSPAN13tetraspanin 13 (217979_at), score: -0.45 TXNthioredoxin (216609_at), score: 0.63 UCP2uncoupling protein 2 (mitochondrial, proton carrier) (208998_at), score: 1 UFM1ubiquitin-fold modifier 1 (218050_at), score: -0.48 VPS11vacuolar protein sorting 11 homolog (S. cerevisiae) (203292_s_at), score: 0.6 XPO4exportin 4 (218479_s_at), score: 0.7 YIPF1Yip1 domain family, member 1 (214733_s_at), score: 0.62 ZC3H4zinc finger CCCH-type containing 4 (213390_at), score: 0.61 ZNF174zinc finger protein 174 (210291_s_at), score: 0.69 ZNF185zinc finger protein 185 (LIM domain) (203585_at), score: -0.57 ZNF219zinc finger protein 219 (219314_s_at), score: -0.49 ZNF22zinc finger protein 22 (KOX 15) (218005_at), score: 0.69 ZNF350zinc finger protein 350 (219266_at), score: 0.67 ZNF434zinc finger protein 434 (218937_at), score: 0.64 ZNF536zinc finger protein 536 (206403_at), score: -0.51 ZNF749zinc finger protein 749 (215289_at), score: -0.57 ZRANB1zinc finger, RAN-binding domain containing 1 (201219_at), score: 0.64
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-3860-raw-cel-1561690199.cel | 1 | 5 | HGPS | hgu133a | none | GM0316B |
| E-GEOD-3407-raw-cel-1437949655.cel | 3 | 4 | Cockayne | hgu133a | none | CSB |
| E-TABM-263-raw-cel-1515486011.cel | 19 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486391.cel | 38 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486191.cel | 28 | 6 | Cycle | hgu133a2 | none | Cycle 1 |