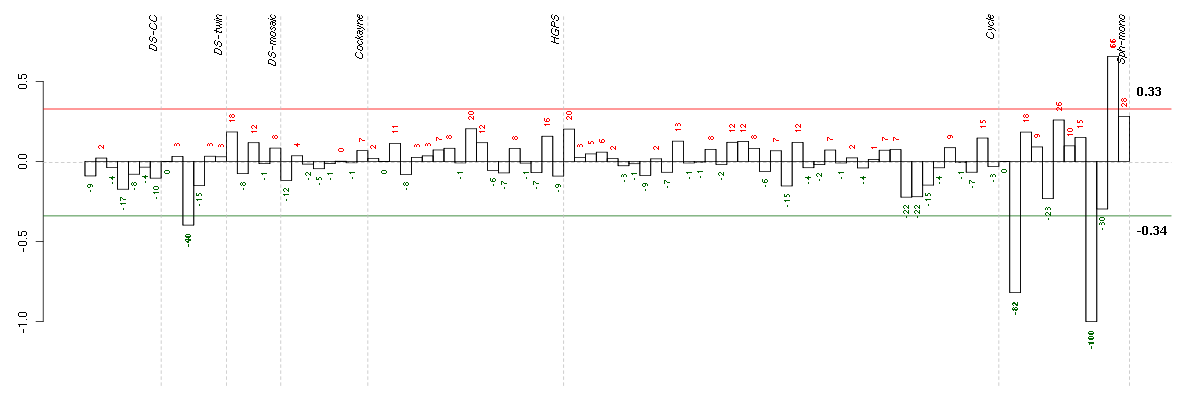

Under-expression is coded with green,
over-expression with red color.



| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 00280 | 3.161e-03 | 1.266 | 8 | 42 | Valine, leucine and isoleucine degradation |
| 00640 | 1.009e-02 | 0.8136 | 6 | 27 | Propanoate metabolism |
| 00010 | 1.407e-02 | 1.235 | 7 | 41 | Glycolysis / Gluconeogenesis |
| 00220 | 2.191e-02 | 0.6328 | 5 | 21 | Urea cycle and metabolism of amino groups |
| 00650 | 4.665e-02 | 0.7835 | 5 | 26 | Butanoate metabolism |
ABCC1ATP-binding cassette, sub-family C (CFTR/MRP), member 1 (202804_at), score: -0.58 ABCC3ATP-binding cassette, sub-family C (CFTR/MRP), member 3 (208161_s_at), score: -0.64 ABCC4ATP-binding cassette, sub-family C (CFTR/MRP), member 4 (203196_at), score: -0.62 ABCG2ATP-binding cassette, sub-family G (WHITE), member 2 (209735_at), score: -0.66 ACSL5acyl-CoA synthetase long-chain family member 5 (218322_s_at), score: -0.8 AENapoptosis enhancing nuclease (219361_s_at), score: 0.66 AHCTF1AT hook containing transcription factor 1 (214766_s_at), score: 0.52 AKR1B10aldo-keto reductase family 1, member B10 (aldose reductase) (206561_s_at), score: -0.89 AKR1C2aldo-keto reductase family 1, member C2 (dihydrodiol dehydrogenase 2; bile acid binding protein; 3-alpha hydroxysteroid dehydrogenase, type III) (209699_x_at), score: -0.6 AKR1C3aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II) (209160_at), score: -0.6 ALDH1A3aldehyde dehydrogenase 1 family, member A3 (203180_at), score: -0.64 ALDH3A1aldehyde dehydrogenase 3 family, memberA1 (205623_at), score: -0.77 ALDH3A2aldehyde dehydrogenase 3 family, member A2 (202053_s_at), score: -0.62 ALDH6A1aldehyde dehydrogenase 6 family, member A1 (221589_s_at), score: -0.66 AMD1adenosylmethionine decarboxylase 1 (201196_s_at), score: 0.6 ANXA4annexin A4 (201301_s_at), score: -0.59 APOBEC3Gapolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3G (204205_at), score: -0.54 APOL2apolipoprotein L, 2 (221653_x_at), score: -0.55 ARMETarginine-rich, mutated in early stage tumors (202655_at), score: -0.6 ASB9ankyrin repeat and SOCS box-containing 9 (205673_s_at), score: -0.67 ATF6Bactivating transcription factor 6 beta (203168_at), score: -0.55 ATRNattractin (212517_at), score: -0.56 B3GNT1UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 1 (203188_at), score: -0.58 BAZ1Abromodomain adjacent to zinc finger domain, 1A (217985_s_at), score: 0.67 BCCIPBRCA2 and CDKN1A interacting protein (218264_at), score: -0.56 BCKDHBbranched chain keto acid dehydrogenase E1, beta polypeptide (210653_s_at), score: -0.6 BET1blocked early in transport 1 homolog (S. cerevisiae) (202710_at), score: -0.56 BLVRBbiliverdin reductase B (flavin reductase (NADPH)) (202201_at), score: -0.58 BMP7bone morphogenetic protein 7 (209590_at), score: -0.7 C11orf71chromosome 11 open reading frame 71 (218789_s_at), score: -0.66 C11orf73chromosome 11 open reading frame 73 (219979_s_at), score: -0.55 C12orf4chromosome 12 open reading frame 4 (218374_s_at), score: 0.54 C12orf41chromosome 12 open reading frame 41 (221821_s_at), score: 0.52 C13orf23chromosome 13 open reading frame 23 (218420_s_at), score: 0.64 C14orf169chromosome 14 open reading frame 169 (219526_at), score: 0.52 C14orf2chromosome 14 open reading frame 2 (202279_at), score: 0.54 C19orf54chromosome 19 open reading frame 54 (222052_at), score: -0.59 C20orf39chromosome 20 open reading frame 39 (219310_at), score: -0.91 C2orf28chromosome 2 open reading frame 28 (219329_s_at), score: -0.56 C3orf64chromosome 3 open reading frame 64 (221935_s_at), score: 0.53 C6orf130chromosome 6 open reading frame 130 (213322_at), score: -0.56 C9orf78chromosome 9 open reading frame 78 (218116_at), score: 0.61 CADPS2Ca++-dependent secretion activator 2 (219572_at), score: -0.65 CBY1chibby homolog 1 (Drosophila) (203450_at), score: -0.7 CCDC86coiled-coil domain containing 86 (203119_at), score: 0.52 CCNJcyclin J (219470_x_at), score: 0.59 CCPG1cell cycle progression 1 (221156_x_at), score: -0.56 CDC2L2cell division cycle 2-like 2 (PITSLRE proteins) (212401_s_at), score: 0.6 CDK5RAP2CDK5 regulatory subunit associated protein 2 (220935_s_at), score: -0.57 CDKN2AIPCDKN2A interacting protein (218929_at), score: 0.53 CEBPZCCAAT/enhancer binding protein (C/EBP), zeta (203341_at), score: 0.6 CELSR1cadherin, EGF LAG seven-pass G-type receptor 1 (flamingo homolog, Drosophila) (41660_at), score: -0.91 CELSR3cadherin, EGF LAG seven-pass G-type receptor 3 (flamingo homolog, Drosophila) (40020_at), score: -0.55 CEP135centrosomal protein 135kDa (206003_at), score: 0.6 CFIcomplement factor I (203854_at), score: -0.68 CHD1chromodomain helicase DNA binding protein 1 (204258_at), score: 0.61 CHFRcheckpoint with forkhead and ring finger domains (218803_at), score: 0.58 CLNS1Achloride channel, nucleotide-sensitive, 1A (209143_s_at), score: -0.56 CPS1carbamoyl-phosphate synthetase 1, mitochondrial (204920_at), score: -0.76 CROTcarnitine O-octanoyltransferase (204573_at), score: -0.65 CRY1cryptochrome 1 (photolyase-like) (209674_at), score: 0.57 CRYZL1crystallin, zeta (quinone reductase)-like 1 (219767_s_at), score: -0.62 CSTBcystatin B (stefin B) (201201_at), score: -0.59 CUTCcutC copper transporter homolog (E. coli) (218970_s_at), score: -0.59 CYB561cytochrome b-561 (209163_at), score: -0.63 DDIT4DNA-damage-inducible transcript 4 (202887_s_at), score: -0.64 DDX39DEAD (Asp-Glu-Ala-Asp) box polypeptide 39 (201584_s_at), score: 0.52 DDX52DEAD (Asp-Glu-Ala-Asp) box polypeptide 52 (212834_at), score: 0.54 DEXIdexamethasone-induced transcript (203733_at), score: 0.54 DHRS12dehydrogenase/reductase (SDR family) member 12 (204800_s_at), score: -0.59 DHX15DEAH (Asp-Glu-Ala-His) box polypeptide 15 (201386_s_at), score: 0.61 DMPKdystrophia myotonica-protein kinase (37996_s_at), score: 0.53 DNAJA1DnaJ (Hsp40) homolog, subfamily A, member 1 (200880_at), score: 0.58 DPH5DPH5 homolog (S. cerevisiae) (219590_x_at), score: -0.62 DPY19L1dpy-19-like 1 (C. elegans) (212792_at), score: -0.62 DUSP14dual specificity phosphatase 14 (203367_at), score: 0.51 EDEM2ER degradation enhancer, mannosidase alpha-like 2 (218282_at), score: -0.55 EGLN3egl nine homolog 3 (C. elegans) (219232_s_at), score: -0.72 EIF5A2eukaryotic translation initiation factor 5A2 (220198_s_at), score: 0.6 ELL3elongation factor RNA polymerase II-like 3 (219517_at), score: -0.56 EML1echinoderm microtubule associated protein like 1 (204797_s_at), score: 0.68 EML3echinoderm microtubule associated protein like 3 (212969_x_at), score: 0.57 EMP1epithelial membrane protein 1 (201325_s_at), score: 0.56 ERMAPerythroblast membrane-associated protein (Scianna blood group) (219905_at), score: -0.61 EXOSC10exosome component 10 (207541_s_at), score: 0.62 FAM131Bfamily with sequence similarity 131, member B (205368_at), score: -0.57 FAM172Afamily with sequence similarity 172, member A (212936_at), score: -0.58 FAM46Afamily with sequence similarity 46, member A (221766_s_at), score: -0.55 FANCFFanconi anemia, complementation group F (218689_at), score: -0.66 FCGR2AFc fragment of IgG, low affinity IIa, receptor (CD32) (203561_at), score: -1 FECHferrochelatase (protoporphyria) (203116_s_at), score: -0.58 FGF13fibroblast growth factor 13 (205110_s_at), score: -0.58 FGFR2fibroblast growth factor receptor 2 (208229_at), score: -0.8 FJX1four jointed box 1 (Drosophila) (219522_at), score: 0.56 FLJ10357hypothetical protein FLJ10357 (58780_s_at), score: 0.56 FLRT1fibronectin leucine rich transmembrane protein 1 (210414_at), score: -0.64 FTLferritin, light polypeptide (213187_x_at), score: -0.68 GABRA2gamma-aminobutyric acid (GABA) A receptor, alpha 2 (207014_at), score: -0.98 GAKcyclin G associated kinase (202281_at), score: 0.57 GAPDHglyceraldehyde-3-phosphate dehydrogenase (AFFX-HUMGAPDH/M33197_M_at), score: -0.58 GBE1glucan (1,4-alpha-), branching enzyme 1 (203282_at), score: -0.62 GEMIN4gem (nuclear organelle) associated protein 4 (205527_s_at), score: 0.53 GRAMD4GRAM domain containing 4 (212856_at), score: -0.59 GREM1gremlin 1, cysteine knot superfamily, homolog (Xenopus laevis) (218469_at), score: 0.76 GYG2glycogenin 2 (215695_s_at), score: -0.6 H2BFSH2B histone family, member S (208579_x_at), score: 0.53 HDAC4histone deacetylase 4 (204225_at), score: -0.59 HIST1H2BKhistone cluster 1, H2bk (209806_at), score: 0.56 HLA-DMAmajor histocompatibility complex, class II, DM alpha (217478_s_at), score: -0.56 HLA-DRB1major histocompatibility complex, class II, DR beta 1 (209312_x_at), score: -0.71 HLA-DRB4major histocompatibility complex, class II, DR beta 4 (208306_x_at), score: -0.67 HLTFhelicase-like transcription factor (202983_at), score: -0.55 HMGXB3HMG box domain containing 3 (212431_at), score: 0.56 HNRNPDheterogeneous nuclear ribonucleoprotein D (AU-rich element RNA binding protein 1, 37kDa) (213359_at), score: 0.51 HSD17B4hydroxysteroid (17-beta) dehydrogenase 4 (201413_at), score: -0.71 HSPA12Aheat shock 70kDa protein 12A (214434_at), score: 0.58 IER5immediate early response 5 (218611_at), score: 0.54 IGSF3immunoglobulin superfamily, member 3 (202421_at), score: -0.64 IQCGIQ motif containing G (221185_s_at), score: -0.62 IRAK1interleukin-1 receptor-associated kinase 1 (201587_s_at), score: 0.72 ITPK1inositol 1,3,4-triphosphate 5/6 kinase (210740_s_at), score: 0.52 ITPR1inositol 1,4,5-triphosphate receptor, type 1 (203710_at), score: -0.59 JTV1JTV1 gene (209972_s_at), score: -0.58 KCNE4potassium voltage-gated channel, Isk-related family, member 4 (222379_at), score: -0.59 KCNMA1potassium large conductance calcium-activated channel, subfamily M, alpha member 1 (221584_s_at), score: -0.55 KIAA0317KIAA0317 (202128_at), score: 0.65 KIF16Bkinesin family member 16B (219570_at), score: -0.71 KLHL18kelch-like 18 (Drosophila) (212882_at), score: 0.56 KYNUkynureninase (L-kynurenine hydrolase) (217388_s_at), score: -0.93 LARSleucyl-tRNA synthetase (217810_x_at), score: -0.56 LBA1lupus brain antigen 1 (213261_at), score: -0.65 LDHAlactate dehydrogenase A (200650_s_at), score: -0.55 LEF1lymphoid enhancer-binding factor 1 (221558_s_at), score: -0.85 LMAN2lectin, mannose-binding 2 (200805_at), score: -0.64 LOC201229hypothetical protein LOC201229 (213195_at), score: -0.77 LOC339047hypothetical protein LOC339047 (221501_x_at), score: 0.55 LOC399491LOC399491 protein (214035_x_at), score: 0.52 LRRFIP2leucine rich repeat (in FLII) interacting protein 2 (220610_s_at), score: 0.53 LUZP1leucine zipper protein 1 (221832_s_at), score: 0.56 LYSTlysosomal trafficking regulator (203518_at), score: -0.61 MAFFv-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian) (36711_at), score: 0.62 MAP2K6mitogen-activated protein kinase kinase 6 (205698_s_at), score: -0.59 MBD1methyl-CpG binding domain protein 1 (203353_s_at), score: 0.59 MCCC2methylcrotonoyl-Coenzyme A carboxylase 2 (beta) (209623_at), score: -0.58 MED20mediator complex subunit 20 (212872_s_at), score: -0.55 MEF2Cmyocyte enhancer factor 2C (209199_s_at), score: 0.52 METT11D1methyltransferase 11 domain containing 1 (218366_x_at), score: 0.61 MPPED2metallophosphoesterase domain containing 2 (205413_at), score: -0.93 MRPS27mitochondrial ribosomal protein S27 (212145_at), score: -0.56 MYO1Bmyosin IB (212364_at), score: 0.61 NAT15N-acetyltransferase 15 (GCN5-related, putative) (45526_g_at), score: 0.57 NBR1neighbor of BRCA1 gene 1 (201384_s_at), score: -0.56 NDRG1N-myc downstream regulated 1 (200632_s_at), score: -0.72 NDUFB5NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5, 16kDa (203621_at), score: -0.62 NFE2L2nuclear factor (erythroid-derived 2)-like 2 (201146_at), score: -0.57 NFE2L3nuclear factor (erythroid-derived 2)-like 3 (204702_s_at), score: -0.57 NOP2NOP2 nucleolar protein homolog (yeast) (214427_at), score: 0.55 NPIPnuclear pore complex interacting protein (204538_x_at), score: 0.52 NPTX1neuronal pentraxin I (204684_at), score: -0.59 NUDT2nudix (nucleoside diphosphate linked moiety X)-type motif 2 (218609_s_at), score: -0.54 NUP214nucleoporin 214kDa (202155_s_at), score: 0.57 NUPL1nucleoporin like 1 (204435_at), score: 0.54 NUPR1nuclear protein 1 (209230_s_at), score: -0.67 NXF1nuclear RNA export factor 1 (208922_s_at), score: 0.61 ORAI3ORAI calcium release-activated calcium modulator 3 (221864_at), score: -0.69 PALM2-AKAP2PALM2-AKAP2 readthrough transcript (202760_s_at), score: 0.54 PANX1pannexin 1 (204715_at), score: 0.52 PCCApropionyl Coenzyme A carboxylase, alpha polypeptide (203860_at), score: -0.56 PCYOX1Lprenylcysteine oxidase 1 like (218953_s_at), score: -0.63 PDGFDplatelet derived growth factor D (219304_s_at), score: -0.7 PEX11Aperoxisomal biogenesis factor 11 alpha (205160_at), score: -0.62 PGDphosphogluconate dehydrogenase (201118_at), score: -0.61 PGK1phosphoglycerate kinase 1 (217356_s_at), score: -0.63 PGM1phosphoglucomutase 1 (201968_s_at), score: -0.63 PHF14PHD finger protein 14 (204525_at), score: -0.55 PIGBphosphatidylinositol glycan anchor biosynthesis, class B (205452_at), score: -0.57 PIGPphosphatidylinositol glycan anchor biosynthesis, class P (221689_s_at), score: -0.68 PIRpirin (iron-binding nuclear protein) (207469_s_at), score: -0.55 PNPOpyridoxamine 5'-phosphate oxidase (218511_s_at), score: -0.63 PPAP2Aphosphatidic acid phosphatase type 2A (209147_s_at), score: -0.6 PPP2CBprotein phosphatase 2 (formerly 2A), catalytic subunit, beta isoform (201375_s_at), score: 0.54 PPP3CCprotein phosphatase 3 (formerly 2B), catalytic subunit, gamma isoform (207000_s_at), score: 0.53 PQLC1PQ loop repeat containing 1 (218208_at), score: 0.53 PRCPprolylcarboxypeptidase (angiotensinase C) (201494_at), score: -0.54 PRMT3protein arginine methyltransferase 3 (213320_at), score: -0.58 PRPF39PRP39 pre-mRNA processing factor 39 homolog (S. cerevisiae) (220553_s_at), score: 0.53 PRRX1paired related homeobox 1 (205991_s_at), score: 0.58 PTPN9protein tyrosine phosphatase, non-receptor type 9 (202958_at), score: 0.56 RAB11FIP1RAB11 family interacting protein 1 (class I) (219681_s_at), score: 0.55 RAG1AP1recombination activating gene 1 activating protein 1 (219125_s_at), score: -0.6 RARBretinoic acid receptor, beta (205080_at), score: -0.82 RARRES1retinoic acid receptor responder (tazarotene induced) 1 (221872_at), score: -0.76 RBL2retinoblastoma-like 2 (p130) (212331_at), score: -0.6 RBM47RNA binding motif protein 47 (218035_s_at), score: -0.77 RIPK4receptor-interacting serine-threonine kinase 4 (221215_s_at), score: -0.57 RNF2ring finger protein 2 (205215_at), score: -0.55 RPIAribose 5-phosphate isomerase A (212973_at), score: 0.61 RTN3reticulon 3 (219549_s_at), score: -0.55 RWDD2BRWD domain containing 2B (218377_s_at), score: -0.62 SAMD4Asterile alpha motif domain containing 4A (212845_at), score: 0.59 SCG5secretogranin V (7B2 protein) (203889_at), score: -0.56 SDHDsuccinate dehydrogenase complex, subunit D, integral membrane protein (202026_at), score: -0.56 SEMA3Csema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C (203789_s_at), score: -0.73 SEPP1selenoprotein P, plasma, 1 (201427_s_at), score: -0.57 SERTAD2SERTA domain containing 2 (202657_s_at), score: 0.56 SF3B2splicing factor 3b, subunit 2, 145kDa (200619_at), score: 0.54 SFRS12splicing factor, arginine/serine-rich 12 (212721_at), score: 0.58 SGCEsarcoglycan, epsilon (204688_at), score: -0.55 SH3BGRSH3 domain binding glutamic acid-rich protein (204979_s_at), score: -0.78 SH3GL1SH3-domain GRB2-like 1 (201851_at), score: 0.65 SLC12A8solute carrier family 12 (potassium/chloride transporters), member 8 (219874_at), score: -0.56 SLC14A1solute carrier family 14 (urea transporter), member 1 (Kidd blood group) (205856_at), score: -0.63 SLC22A5solute carrier family 22 (organic cation/carnitine transporter), member 5 (205074_at), score: -0.66 SLC4A7solute carrier family 4, sodium bicarbonate cotransporter, member 7 (209884_s_at), score: -0.66 SMAD7SMAD family member 7 (204790_at), score: 0.52 SNAP25synaptosomal-associated protein, 25kDa (202508_s_at), score: -0.59 SNCAIPsynuclein, alpha interacting protein (219511_s_at), score: -0.69 SOCS2suppressor of cytokine signaling 2 (203373_at), score: 0.54 SPAG16sperm associated antigen 16 (219109_at), score: -0.6 SPANXCSPANX family, member C (220217_x_at), score: -0.63 SPCS2signal peptidase complex subunit 2 homolog (S. cerevisiae) (201239_s_at), score: -0.57 SPRY2sprouty homolog 2 (Drosophila) (204011_at), score: 0.58 STAG1stromal antigen 1 (202293_at), score: 0.71 SULT1A2sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2 (207122_x_at), score: -0.6 TACC2transforming, acidic coiled-coil containing protein 2 (202289_s_at), score: 0.62 TBC1D12TBC1 domain family, member 12 (221858_at), score: 0.53 TCP1t-complex 1 (208778_s_at), score: 0.58 TGFBItransforming growth factor, beta-induced, 68kDa (201506_at), score: -0.55 THEM2thioesterase superfamily member 2 (204565_at), score: -0.64 THNSL1threonine synthase-like 1 (S. cerevisiae) (219800_s_at), score: -0.67 TIAL1TIA1 cytotoxic granule-associated RNA binding protein-like 1 (202406_s_at), score: 0.55 TK2thymidine kinase 2, mitochondrial (204276_at), score: -0.56 TMEM100transmembrane protein 100 (219230_at), score: -0.57 TMEM50Btransmembrane protein 50B (219600_s_at), score: -0.55 TP53TG1TP53 target 1 (non-protein coding) (209917_s_at), score: -0.57 TRAPPC6Atrafficking protein particle complex 6A (204985_s_at), score: -0.62 TSC22D3TSC22 domain family, member 3 (208763_s_at), score: -0.58 TSPAN31tetraspanin 31 (203227_s_at), score: -0.55 TTC30Atetratricopeptide repeat domain 30A (213679_at), score: -0.64 TTC35tetratricopeptide repeat domain 35 (203584_at), score: -0.59 TWISTNBTWIST neighbor (214729_at), score: 0.67 TYK2tyrosine kinase 2 (205546_s_at), score: 0.58 UBE2Kubiquitin-conjugating enzyme E2K (UBC1 homolog, yeast) (202347_s_at), score: 0.54 UBE2L6ubiquitin-conjugating enzyme E2L 6 (201649_at), score: -0.61 UBR7ubiquitin protein ligase E3 component n-recognin 7 (putative) (218108_at), score: 0.52 UPP1uridine phosphorylase 1 (203234_at), score: 0.53 UQCRC2ubiquinol-cytochrome c reductase core protein II (200883_at), score: -0.55 UTP3UTP3, small subunit (SSU) processome component, homolog (S. cerevisiae) (209486_at), score: 0.56 VAV3vav 3 guanine nucleotide exchange factor (218807_at), score: -0.71 VCAM1vascular cell adhesion molecule 1 (203868_s_at), score: -0.81 VPS33Avacuolar protein sorting 33 homolog A (S. cerevisiae) (204590_x_at), score: 0.56 WDR37WD repeat domain 37 (211383_s_at), score: 0.57 WDR43WD repeat domain 43 (214662_at), score: 0.56 WDR5BWD repeat domain 5B (219538_at), score: -0.66 YAF2YY1 associated factor 2 (206238_s_at), score: 0.53 YIPF6Yip1 domain family, member 6 (212341_at), score: -0.57 ZC3H14zinc finger CCCH-type containing 14 (204216_s_at), score: 0.67 ZNF143zinc finger protein 143 (221873_at), score: 0.64 ZNF189zinc finger protein 189 (207513_s_at), score: -0.55 ZNF248zinc finger protein 248 (213269_at), score: -0.75 ZNF323zinc finger protein 323 (222016_s_at), score: -0.6 ZNF592zinc finger protein 592 (204473_s_at), score: 0.59 ZNF629zinc finger protein 629 (213196_at), score: 0.52 ZNF747zinc finger protein 747 (206180_x_at), score: -0.59
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-4219-raw-cel-1311956457.cel | 14 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956138.cel | 4 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| 3Twin.CEL | 3 | 2 | DS-twin | hgu133plus2 | Down | DS-twin 3 |
| E-GEOD-4219-raw-cel-1311956634.cel | 19 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |