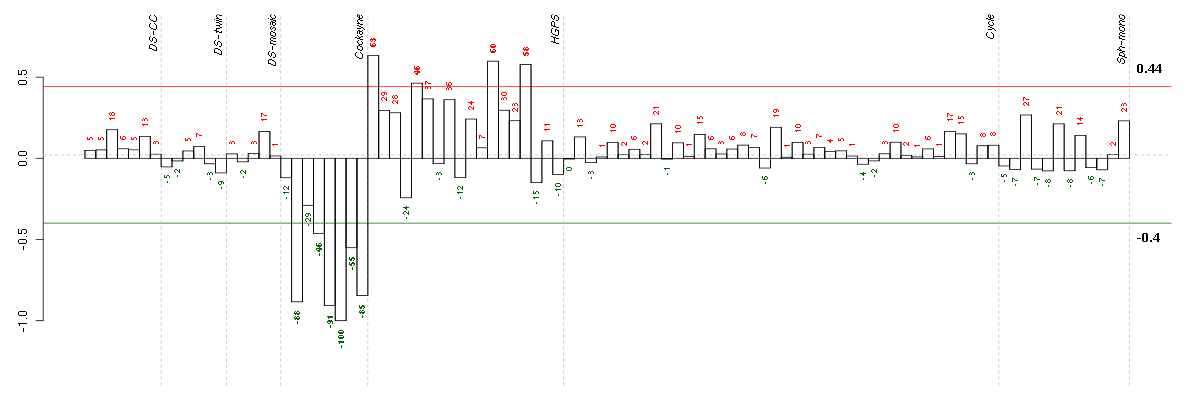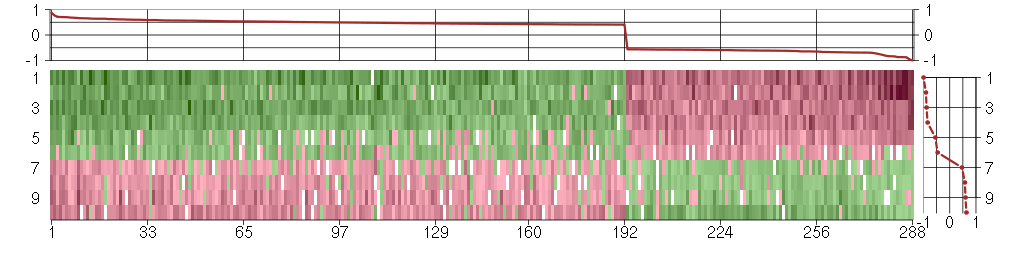

Under-expression is coded with green,
over-expression with red color.


extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
all
This term is the most general term possible

ABCB1ATP-binding cassette, sub-family B (MDR/TAP), member 1 (209993_at), score: -1 ABHD14Aabhydrolase domain containing 14A (210006_at), score: 0.51 ACIN1apoptotic chromatin condensation inducer 1 (201715_s_at), score: -0.61 ACP2acid phosphatase 2, lysosomal (202767_at), score: 0.42 ACSL3acyl-CoA synthetase long-chain family member 3 (201660_at), score: 0.43 ACTC1actin, alpha, cardiac muscle 1 (205132_at), score: 0.55 ADORA1adenosine A1 receptor (216220_s_at), score: 0.49 AHNAK2AHNAK nucleoprotein 2 (212992_at), score: 0.47 AIM2absent in melanoma 2 (206513_at), score: -0.69 AKAP8LA kinase (PRKA) anchor protein 8-like (218064_s_at), score: -0.56 ALDOCaldolase C, fructose-bisphosphate (202022_at), score: 0.42 ANK3ankyrin 3, node of Ranvier (ankyrin G) (206385_s_at), score: -0.58 ANP32Aacidic (leucine-rich) nuclear phosphoprotein 32 family, member A (201043_s_at), score: -0.58 APCadenomatous polyposis coli (203525_s_at), score: 0.42 APOL3apolipoprotein L, 3 (221087_s_at), score: -0.67 ARHGAP1Rho GTPase activating protein 1 (202117_at), score: 0.42 ARNTLaryl hydrocarbon receptor nuclear translocator-like (209824_s_at), score: 0.52 ARSAarylsulfatase A (204443_at), score: 0.43 ATP6V1B2ATPase, H+ transporting, lysosomal 56/58kDa, V1 subunit B2 (201089_at), score: -0.56 ATP9AATPase, class II, type 9A (212062_at), score: 0.41 BAALCbrain and acute leukemia, cytoplasmic (218899_s_at), score: 0.57 BAZ2Abromodomain adjacent to zinc finger domain, 2A (201353_s_at), score: -0.73 BCKDKbranched chain ketoacid dehydrogenase kinase (202030_at), score: 0.49 BHMT2betaine-homocysteine methyltransferase 2 (219902_at), score: -0.66 C10orf116chromosome 10 open reading frame 116 (203571_s_at), score: 0.65 C17orf70chromosome 17 open reading frame 70 (221800_s_at), score: 0.46 C20orf149chromosome 20 open reading frame 149 (218010_x_at), score: 0.55 C3orf64chromosome 3 open reading frame 64 (221935_s_at), score: 0.59 C4orf31chromosome 4 open reading frame 31 (219747_at), score: 0.87 C5orf23chromosome 5 open reading frame 23 (219054_at), score: 0.57 C5orf30chromosome 5 open reading frame 30 (221823_at), score: 0.64 CAPGcapping protein (actin filament), gelsolin-like (201850_at), score: 0.54 CARD10caspase recruitment domain family, member 10 (210026_s_at), score: -0.6 CD248CD248 molecule, endosialin (219025_at), score: 0.45 CD81CD81 molecule (200675_at), score: 0.44 CDH4cadherin 4, type 1, R-cadherin (retinal) (220227_at), score: 0.41 CDKN1Acyclin-dependent kinase inhibitor 1A (p21, Cip1) (202284_s_at), score: 0.41 CDR2Lcerebellar degeneration-related protein 2-like (213230_at), score: 0.62 CGBchorionic gonadotropin, beta polypeptide (205387_s_at), score: 0.66 CH25Hcholesterol 25-hydroxylase (206932_at), score: 0.5 CHMchoroideremia (Rab escort protein 1) (207099_s_at), score: -0.58 CHPFchondroitin polymerizing factor (202175_at), score: 0.48 CKBcreatine kinase, brain (200884_at), score: 0.54 CLINT1clathrin interactor 1 (201768_s_at), score: -0.59 COL11A1collagen, type XI, alpha 1 (37892_at), score: 0.5 COL8A2collagen, type VIII, alpha 2 (221900_at), score: 0.57 COMPcartilage oligomeric matrix protein (205713_s_at), score: 0.76 COPZ2coatomer protein complex, subunit zeta 2 (219561_at), score: 0.47 CRATcarnitine acetyltransferase (209522_s_at), score: 0.53 CRTAPcartilage associated protein (201380_at), score: 0.48 CRYABcrystallin, alpha B (209283_at), score: 0.57 CTBP1C-terminal binding protein 1 (213979_s_at), score: -0.61 CXorf15chromosome X open reading frame 15 (219969_at), score: -0.57 CYBAcytochrome b-245, alpha polypeptide (203028_s_at), score: 0.49 DDX17DEAD (Asp-Glu-Ala-Asp) box polypeptide 17 (208151_x_at), score: -0.84 DDX28DEAD (Asp-Glu-Ala-Asp) box polypeptide 28 (203785_s_at), score: -0.59 DDX3XDEAD (Asp-Glu-Ala-Asp) box polypeptide 3, X-linked (201211_s_at), score: -0.67 DHX9DEAH (Asp-Glu-Ala-His) box polypeptide 9 (212105_s_at), score: -0.57 DND1dead end homolog 1 (zebrafish) (57739_at), score: -0.6 DPYSL4dihydropyrimidinase-like 4 (205493_s_at), score: 0.44 DUSP14dual specificity phosphatase 14 (203367_at), score: 0.52 ECM1extracellular matrix protein 1 (209365_s_at), score: 0.52 EIF1AYeukaryotic translation initiation factor 1A, Y-linked (204409_s_at), score: 0.51 EPB41L3erythrocyte membrane protein band 4.1-like 3 (206710_s_at), score: 0.49 EPHB2EPH receptor B2 (209589_s_at), score: -0.57 FAM108A1family with sequence similarity 108, member A1 (221267_s_at), score: 0.61 FAM155Afamily with sequence similarity 155, member A (214825_at), score: 0.43 FAM169Afamily with sequence similarity 169, member A (213954_at), score: -0.69 FASFas (TNF receptor superfamily, member 6) (204780_s_at), score: 0.53 FAT1FAT tumor suppressor homolog 1 (Drosophila) (201579_at), score: 0.56 FBN2fibrillin 2 (203184_at), score: 0.52 FBXW7F-box and WD repeat domain containing 7 (218751_s_at), score: 0.5 FHOD3formin homology 2 domain containing 3 (218980_at), score: 0.47 FKBP4FK506 binding protein 4, 59kDa (200894_s_at), score: -0.62 FKBP9FK506 binding protein 9, 63 kDa (212169_at), score: 0.64 FLRT2fibronectin leucine rich transmembrane protein 2 (204359_at), score: 0.43 FOXD1forkhead box D1 (206307_s_at), score: 0.41 GABBR2gamma-aminobutyric acid (GABA) B receptor, 2 (209990_s_at), score: 0.58 GAGE4G antigen 4 (207086_x_at), score: -0.87 GAGE6G antigen 6 (208155_x_at), score: -0.87 GAS6growth arrest-specific 6 (202177_at), score: 0.56 GATA3GATA binding protein 3 (209604_s_at), score: -0.75 GBP1guanylate binding protein 1, interferon-inducible, 67kDa (202269_x_at), score: -0.63 GLSglutaminase (203159_at), score: 0.59 GLT25D2glycosyltransferase 25 domain containing 2 (209883_at), score: 0.46 GLTPglycolipid transfer protein (219267_at), score: 0.56 GMDSGDP-mannose 4,6-dehydratase (204875_s_at), score: -0.57 GYS1glycogen synthase 1 (muscle) (201673_s_at), score: 0.52 HAS2hyaluronan synthase 2 (206432_at), score: 0.55 HEPHhephaestin (203903_s_at), score: 0.43 HERC5hect domain and RLD 5 (219863_at), score: -0.71 HGSNATheparan-alpha-glucosaminide N-acetyltransferase (218017_s_at), score: 0.48 HIPK1homeodomain interacting protein kinase 1 (212291_at), score: -0.62 HIPK2homeodomain interacting protein kinase 2 (219028_at), score: -0.6 HIST1H4Chistone cluster 1, H4c (205967_at), score: 0.56 HIST2H2AA3histone cluster 2, H2aa3 (214290_s_at), score: 0.53 HIVEP2human immunodeficiency virus type I enhancer binding protein 2 (212642_s_at), score: 0.55 HMOX1heme oxygenase (decycling) 1 (203665_at), score: 0.45 HOXA10homeobox A10 (213150_at), score: 0.47 HOXA9homeobox A9 (214651_s_at), score: 0.43 HS3ST3A1heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1 (219985_at), score: 0.41 HSPA4Lheat shock 70kDa protein 4-like (205543_at), score: -0.65 HSPB2heat shock 27kDa protein 2 (205824_at), score: 0.54 HSPB6heat shock protein, alpha-crystallin-related, B6 (214767_s_at), score: 0.42 HSPB7heat shock 27kDa protein family, member 7 (cardiovascular) (218934_s_at), score: 0.71 IFI16interferon, gamma-inducible protein 16 (208965_s_at), score: -0.68 IFIT3interferon-induced protein with tetratricopeptide repeats 3 (204747_at), score: -0.59 IFRD1interferon-related developmental regulator 1 (202147_s_at), score: -0.56 IGFBP3insulin-like growth factor binding protein 3 (212143_s_at), score: 0.64 IL21Rinterleukin 21 receptor (219971_at), score: 0.41 INF2inverted formin, FH2 and WH2 domain containing (218144_s_at), score: 0.45 INHBBinhibin, beta B (205258_at), score: 0.53 IPO8importin 8 (205701_at), score: -0.65 IRS2insulin receptor substrate 2 (209185_s_at), score: 0.64 IRX5iroquois homeobox 5 (210239_at), score: 0.59 ISLRimmunoglobulin superfamily containing leucine-rich repeat (207191_s_at), score: 0.7 ITGBL1integrin, beta-like 1 (with EGF-like repeat domains) (214927_at), score: 0.42 JARID1Djumonji, AT rich interactive domain 1D (206700_s_at), score: 0.41 KBTBD11kelch repeat and BTB (POZ) domain containing 11 (204301_at), score: 0.45 KIAA0495KIAA0495 (213340_s_at), score: 0.62 KIAA1539KIAA1539 (207765_s_at), score: 0.44 KIF3Akinesin family member 3A (213623_at), score: -0.63 KRT14keratin 14 (209351_at), score: 0.56 KRT19keratin 19 (201650_at), score: 0.6 LAMB3laminin, beta 3 (209270_at), score: -0.61 LAPTM5lysosomal multispanning membrane protein 5 (201721_s_at), score: 0.44 LBHlimb bud and heart development homolog (mouse) (221011_s_at), score: 0.54 LDOC1leucine zipper, down-regulated in cancer 1 (204454_at), score: 0.44 LEF1lymphoid enhancer-binding factor 1 (221558_s_at), score: -0.56 LIPAlipase A, lysosomal acid, cholesterol esterase (201847_at), score: 0.44 LMF1lipase maturation factor 1 (219135_s_at), score: 0.49 LOC151162hypothetical LOC151162 (212098_at), score: 0.42 LOC388796hypothetical LOC388796 (65588_at), score: -0.61 LOC728855hypothetical LOC728855 (222001_x_at), score: -0.64 LOC730092RRN3 RNA polymerase I transcription factor homolog (S. cerevisiae) pseudogene (216908_x_at), score: -0.58 LRIG2leucine-rich repeats and immunoglobulin-like domains 2 (205953_at), score: -0.57 LRP3low density lipoprotein receptor-related protein 3 (204381_at), score: 0.43 LRRC15leucine rich repeat containing 15 (213909_at), score: 0.47 LRRFIP1leucine rich repeat (in FLII) interacting protein 1 (201861_s_at), score: 0.56 LTBP2latent transforming growth factor beta binding protein 2 (204682_at), score: 0.49 M6PRmannose-6-phosphate receptor (cation dependent) (200900_s_at), score: -0.69 MALLmal, T-cell differentiation protein-like (209373_at), score: 0.53 MAN2A1mannosidase, alpha, class 2A, member 1 (205105_at), score: 0.49 MARCH2membrane-associated ring finger (C3HC4) 2 (210075_at), score: 0.44 MARK1MAP/microtubule affinity-regulating kinase 1 (221047_s_at), score: -0.83 MBOAT2membrane bound O-acyltransferase domain containing 2 (213288_at), score: 0.49 MED6mediator complex subunit 6 (207079_s_at), score: -0.58 MEGF6multiple EGF-like-domains 6 (213942_at), score: 0.51 MFAP5microfibrillar associated protein 5 (213764_s_at), score: 0.6 MITFmicrophthalmia-associated transcription factor (207233_s_at), score: -0.67 MREGmelanoregulin (219648_at), score: 0.42 MRFAP1L1Morf4 family associated protein 1-like 1 (212199_at), score: -0.56 MSCmusculin (activated B-cell factor-1) (209928_s_at), score: -0.63 MT1Gmetallothionein 1G (204745_x_at), score: 0.41 MT1Mmetallothionein 1M (217546_at), score: 0.64 MT1P3metallothionein 1 pseudogene 3 (221953_s_at), score: 0.55 MT1Xmetallothionein 1X (204326_x_at), score: 0.46 MXRA5matrix-remodelling associated 5 (209596_at), score: 0.42 MXRA8matrix-remodelling associated 8 (213422_s_at), score: 0.5 MYO10myosin X (201976_s_at), score: 0.46 MYO1Cmyosin IC (214656_x_at), score: 0.69 NAPGN-ethylmaleimide-sensitive factor attachment protein, gamma (210048_at), score: -0.62 NBL1neuroblastoma, suppression of tumorigenicity 1 (37005_at), score: 0.42 NCR2natural cytotoxicity triggering receptor 2 (217045_x_at), score: 0.5 NCRNA00084non-protein coding RNA 84 (214657_s_at), score: -0.6 NDNnecdin homolog (mouse) (209550_at), score: 0.58 NDRG3NDRG family member 3 (217286_s_at), score: 0.41 NEFLneurofilament, light polypeptide (221805_at), score: -0.68 NEK7NIMA (never in mitosis gene a)-related kinase 7 (212530_at), score: 0.48 NFAT5nuclear factor of activated T-cells 5, tonicity-responsive (208003_s_at), score: -0.58 NGDNneuroguidin, EIF4E binding protein (213794_s_at), score: -0.61 NMUneuromedin U (206023_at), score: -0.61 NONOnon-POU domain containing, octamer-binding (208698_s_at), score: -0.57 NPDC1neural proliferation, differentiation and control, 1 (218086_at), score: 0.43 NRG1neuregulin 1 (206343_s_at), score: 0.48 NRGNneurogranin (protein kinase C substrate, RC3) (204081_at), score: -0.62 NTF3neurotrophin 3 (206706_at), score: 0.53 OASL2'-5'-oligoadenylate synthetase-like (210797_s_at), score: -0.96 ODZ4odz, odd Oz/ten-m homolog 4 (Drosophila) (213273_at), score: 0.52 OPCMLopioid binding protein/cell adhesion molecule-like (214111_at), score: 0.53 PARP12poly (ADP-ribose) polymerase family, member 12 (218543_s_at), score: -0.57 PBX1pre-B-cell leukemia homeobox 1 (212148_at), score: 0.57 PCBP4poly(rC) binding protein 4 (209361_s_at), score: 0.43 PDE10Aphosphodiesterase 10A (205501_at), score: -0.68 PDPK13-phosphoinositide dependent protein kinase-1 (204524_at), score: 0.54 PILRBpaired immunoglobin-like type 2 receptor beta (220954_s_at), score: -0.69 PITPNC1phosphatidylinositol transfer protein, cytoplasmic 1 (219155_at), score: 0.41 PLSCR3phospholipid scramblase 3 (218828_at), score: 0.41 PNMA2paraneoplastic antigen MA2 (209598_at), score: -0.62 PODXLpodocalyxin-like (201578_at), score: 0.6 PPM2Cprotein phosphatase 2C, magnesium-dependent, catalytic subunit (218273_s_at), score: -0.64 PPP1R3Cprotein phosphatase 1, regulatory (inhibitor) subunit 3C (204284_at), score: 0.51 PRKCHprotein kinase C, eta (218764_at), score: -0.57 PRPF38BPRP38 pre-mRNA processing factor 38 (yeast) domain containing B (218040_at), score: -0.59 PSG2pregnancy specific beta-1-glycoprotein 2 (208134_x_at), score: 0.49 PSG5pregnancy specific beta-1-glycoprotein 5 (204830_x_at), score: 0.42 PSG9pregnancy specific beta-1-glycoprotein 9 (209594_x_at), score: 0.46 PSMB4proteasome (prosome, macropain) subunit, beta type, 4 (202243_s_at), score: 0.59 PSMB8proteasome (prosome, macropain) subunit, beta type, 8 (large multifunctional peptidase 7) (209040_s_at), score: -0.59 PTGER3prostaglandin E receptor 3 (subtype EP3) (213933_at), score: 0.48 PTHLHparathyroid hormone-like hormone (211756_at), score: 0.41 PTPRBprotein tyrosine phosphatase, receptor type, B (205846_at), score: 0.71 PTPRRprotein tyrosine phosphatase, receptor type, R (210675_s_at), score: 0.52 PXDNperoxidasin homolog (Drosophila) (212013_at), score: 0.42 QPCTglutaminyl-peptide cyclotransferase (205174_s_at), score: 0.43 R3HDM2R3H domain containing 2 (203831_at), score: 0.41 RAB40BRAB40B, member RAS oncogene family (204547_at), score: 0.49 RAB7L1RAB7, member RAS oncogene family-like 1 (218700_s_at), score: -0.69 RABGAP1LRAB GTPase activating protein 1-like (213982_s_at), score: 0.45 RAD23BRAD23 homolog B (S. cerevisiae) (201223_s_at), score: 0.68 RASIP1Ras interacting protein 1 (220027_s_at), score: 0.51 RBM12BRNA binding motif protein 12B (51228_at), score: -0.59 RBM3RNA binding motif (RNP1, RRM) protein 3 (222026_at), score: 0.56 RCBTB2regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 (204759_at), score: 0.42 RDXradixin (204969_s_at), score: -0.59 RFNGRFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase (212968_at), score: 0.42 RHODras homolog gene family, member D (31846_at), score: 0.48 RNF19Aring finger protein 19A (220483_s_at), score: -0.58 RPP14ribonuclease P/MRP 14kDa subunit (204245_s_at), score: -0.58 RPS4Y1ribosomal protein S4, Y-linked 1 (201909_at), score: 0.41 RTN3reticulon 3 (219549_s_at), score: 0.43 SAMD4Asterile alpha motif domain containing 4A (212845_at), score: 0.5 SAP18Sin3A-associated protein, 18kDa (208740_at), score: 0.43 SART3squamous cell carcinoma antigen recognized by T cells 3 (209127_s_at), score: -0.61 SCG5secretogranin V (7B2 protein) (203889_at), score: 0.6 SDC4syndecan 4 (202071_at), score: 0.41 SERPINB7serpin peptidase inhibitor, clade B (ovalbumin), member 7 (206421_s_at), score: 0.44 SERPINE2serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 (212190_at), score: 0.45 SGCDsarcoglycan, delta (35kDa dystrophin-associated glycoprotein) (213543_at), score: 0.62 SIRPAsignal-regulatory protein alpha (202897_at), score: 0.46 SLC10A3solute carrier family 10 (sodium/bile acid cotransporter family), member 3 (204928_s_at), score: 0.5 SLC16A1solute carrier family 16, member 1 (monocarboxylic acid transporter 1) (209900_s_at), score: 0.6 SLC16A5solute carrier family 16, member 5 (monocarboxylic acid transporter 6) (206600_s_at), score: 0.48 SLC2A10solute carrier family 2 (facilitated glucose transporter), member 10 (221024_s_at), score: 0.42 SLC30A1solute carrier family 30 (zinc transporter), member 1 (212907_at), score: 0.59 SLC4A4solute carrier family 4, sodium bicarbonate cotransporter, member 4 (203908_at), score: 0.62 SLC7A1solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 (212295_s_at), score: 0.52 SLC7A11solute carrier family 7, (cationic amino acid transporter, y+ system) member 11 (217678_at), score: -0.68 SMAD5SMAD family member 5 (205187_at), score: -0.57 SMEK1SMEK homolog 1, suppressor of mek1 (Dictyostelium) (220368_s_at), score: -0.64 SMG1SMG1 homolog, phosphatidylinositol 3-kinase-related kinase (C. elegans) (210057_at), score: -0.67 SMPD1sphingomyelin phosphodiesterase 1, acid lysosomal (209420_s_at), score: 0.44 SNCAsynuclein, alpha (non A4 component of amyloid precursor) (204466_s_at), score: -0.78 SSR3signal sequence receptor, gamma (translocon-associated protein gamma) (217790_s_at), score: 0.64 SYTL2synaptotagmin-like 2 (220613_s_at), score: -0.69 TACSTD2tumor-associated calcium signal transducer 2 (202286_s_at), score: 0.61 TBPL1TBP-like 1 (208398_s_at), score: -0.57 TBX5T-box 5 (207155_at), score: 0.47 TCEAL2transcription elongation factor A (SII)-like 2 (211276_at), score: -0.81 TERF1telomeric repeat binding factor (NIMA-interacting) 1 (203448_s_at), score: -0.65 TESK1testis-specific kinase 1 (204106_at), score: 0.44 TGIF1TGFB-induced factor homeobox 1 (203313_s_at), score: -0.58 THNSL2threonine synthase-like 2 (S. cerevisiae) (219044_at), score: 0.43 TIMP2TIMP metallopeptidase inhibitor 2 (203167_at), score: 0.7 TMEM204transmembrane protein 204 (219315_s_at), score: 0.55 TMEM47transmembrane protein 47 (209656_s_at), score: 0.51 TMEM51transmembrane protein 51 (218815_s_at), score: -0.68 TMEM8transmembrane protein 8 (five membrane-spanning domains) (221882_s_at), score: 0.53 TMEM87Atransmembrane protein 87A (212204_at), score: 0.41 TMOD1tropomodulin 1 (203661_s_at), score: -0.69 TOB1transducer of ERBB2, 1 (202704_at), score: 0.57 TRIM16tripartite motif-containing 16 (204341_at), score: -0.6 TSPAN13tetraspanin 13 (217979_at), score: 0.65 TSPYL5TSPY-like 5 (213122_at), score: 0.57 UAP1UDP-N-acteylglucosamine pyrophosphorylase 1 (209340_at), score: 0.67 UBE2D4ubiquitin-conjugating enzyme E2D 4 (putative) (218837_s_at), score: 0.46 UBE2L6ubiquitin-conjugating enzyme E2L 6 (201649_at), score: -0.61 UBL3ubiquitin-like 3 (201535_at), score: 0.66 UGCGUDP-glucose ceramide glucosyltransferase (204881_s_at), score: -0.61 UGT1A1UDP glucuronosyltransferase 1 family, polypeptide A1 (207126_x_at), score: -0.83 UGT1A6UDP glucuronosyltransferase 1 family, polypeptide A6 (206094_x_at), score: -0.87 UGT1A9UDP glucuronosyltransferase 1 family, polypeptide A9 (204532_x_at), score: -0.87 UROSuroporphyrinogen III synthase (203031_s_at), score: 0.47 USP18ubiquitin specific peptidase 18 (219211_at), score: -0.64 VKORC1vitamin K epoxide reductase complex, subunit 1 (217949_s_at), score: 0.41 VLDLRvery low density lipoprotein receptor (209822_s_at), score: 0.41 VWA5Avon Willebrand factor A domain containing 5A (210102_at), score: -0.59 WDR91WD repeat domain 91 (218971_s_at), score: -0.58 WNT5Bwingless-type MMTV integration site family, member 5B (221029_s_at), score: 0.53 WT1Wilms tumor 1 (206067_s_at), score: -0.58 XYLT1xylosyltransferase I (213725_x_at), score: 0.46 YIPF2Yip1 domain family, member 2 (65086_at), score: 0.45 ZNF185zinc finger protein 185 (LIM domain) (203585_at), score: 0.53 ZNF224zinc finger protein 224 (216983_s_at), score: -0.59 ZNF227zinc finger protein 227 (217403_s_at), score: -0.61 ZNF281zinc finger protein 281 (218401_s_at), score: 0.5 ZNF415zinc finger protein 415 (205514_at), score: 0.46 ZNF654zinc finger protein 654 (219239_s_at), score: 0.45
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-3407-raw-cel-1437949750.cel | 6 | 4 | Cockayne | hgu133a | CS | CSB |
| E-GEOD-3407-raw-cel-1437949721.cel | 5 | 4 | Cockayne | hgu133a | CS | CSB |
| E-GEOD-3407-raw-cel-1437949579.cel | 2 | 4 | Cockayne | hgu133a | none | CSB |
| E-GEOD-3407-raw-cel-1437949938.cel | 8 | 4 | Cockayne | hgu133a | none | CSB |
| E-GEOD-3407-raw-cel-1437949854.cel | 7 | 4 | Cockayne | hgu133a | CS | eGFP |
| E-GEOD-3407-raw-cel-1437949704.cel | 4 | 4 | Cockayne | hgu133a | CS | eGFP |
| E-GEOD-3860-raw-cel-1561690248.cel | 5 | 5 | HGPS | hgu133a | HGPS | AG11513 |
| E-GEOD-3860-raw-cel-1561690416.cel | 15 | 5 | HGPS | hgu133a | none | GM0316B |
| E-GEOD-3860-raw-cel-1561690360.cel | 12 | 5 | HGPS | hgu133a | none | GM0316B |
| E-GEOD-3860-raw-cel-1561690199.cel | 1 | 5 | HGPS | hgu133a | none | GM0316B |