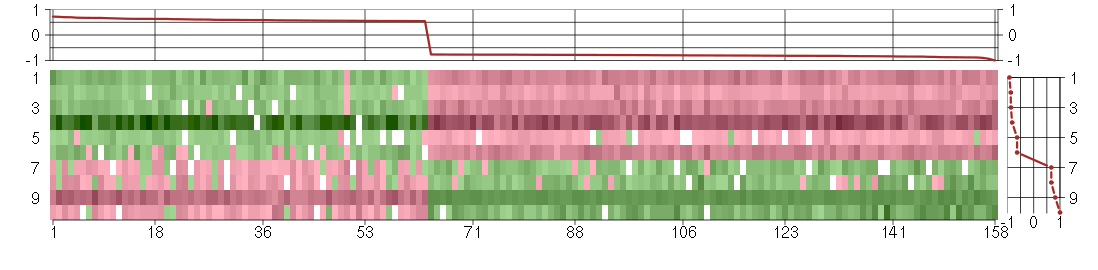

Under-expression is coded with green,
over-expression with red color.

immune system process
Any process involved in the development or functioning of the immune system, an organismal system for calibrated responses to potential internal or invasive threats.
antigen processing and presentation of peptide antigen via MHC class I
The process by which an antigen-presenting cell expresses a peptide antigen on its cell surface in association with an MHC class I protein complex. Class I here refers to classical class I molecules.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
antigen processing and presentation
The process by which an antigen-presenting cell expresses antigen (peptide or lipid) on its cell surface in association with an MHC protein complex.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
antigen processing and presentation of peptide antigen
The process by which an antigen-presenting cell expresses peptide antigen in association with an MHC protein complex on its cell surface, including proteolysis and transport steps for the peptide antigen both prior to and following assembly with the MHC protein complex. The peptide antigen is typically, but not always, processed from an endogenous or exogenous protein.
response to stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
all
This term is the most general term possible

plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
MHC class I protein complex
A transmembrane protein complex composed of a MHC class I alpha chain and an invariant beta2-microglobin chain, and with or without a bound peptide antigen. Class I here refers to classical class I molecules.
extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
extracellular space
That part of a multicellular organism outside the cells proper, usually taken to be outside the plasma membranes, and occupied by fluid.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
macromolecular complex
A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which the constituent parts function together.
MHC protein complex
A transmembrane protein complex composed of an MHC alpha chain and, in most cases, either an MHC class II beta chain or an invariant beta2-microglobin chain, and with or without a bound peptide, lipid, or polysaccharide antigen.
protein complex
Any macromolecular complex composed of two or more polypeptide subunits, which may or may not be identical. Protein complexes may have other associated non-protein prosthetic groups, such as nucleotides, metal ions or carbohydrate groups.
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
MHC protein complex
A transmembrane protein complex composed of an MHC alpha chain and, in most cases, either an MHC class II beta chain or an invariant beta2-microglobin chain, and with or without a bound peptide, lipid, or polysaccharide antigen.

molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
signal transducer activity
Mediates the transfer of a signal from the outside to the inside of a cell by means other than the introduction of the signal molecule itself into the cell.
receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity.
transmembrane receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity, and spanning to the membrane of either the cell or an organelle.
MHC class I receptor activity
Combining with an MHC class I protein complex to initiate a change in cellular activity. Class I here refers to classical class I molecules.
molecular transducer activity
The molecular function that accepts an input of one form and creates an output of a different form.
all
This term is the most general term possible
| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 04514 | 6.523e-03 | 1.432 | 8 | 70 | Cell adhesion molecules (CAMs) |
| 05330 | 4.085e-02 | 0.4297 | 4 | 21 | Allograft rejection |
| 05332 | 4.085e-02 | 0.4297 | 4 | 21 | Graft-versus-host disease |
| 05320 | 4.651e-02 | 0.4501 | 4 | 22 | Autoimmune thyroid disease |
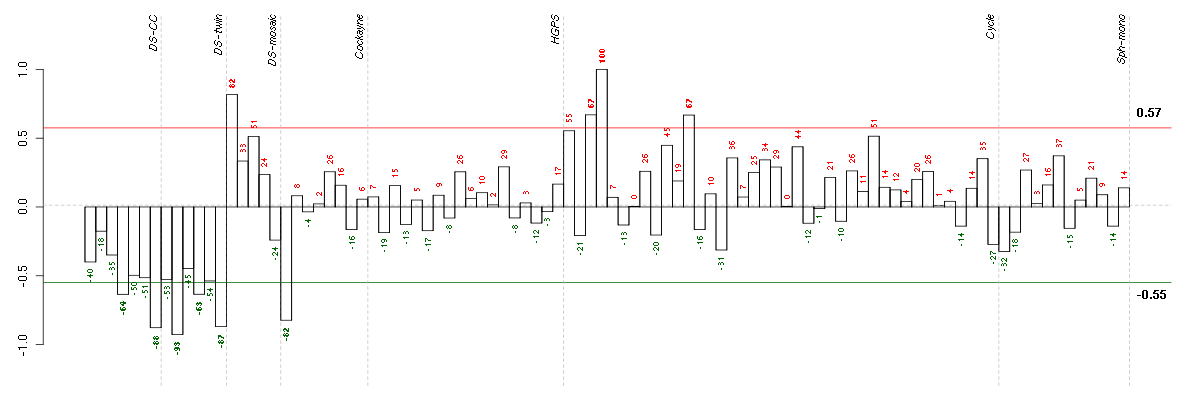
A1CFAPOBEC1 complementation factor (220951_s_at), score: -0.79 ABCG4ATP-binding cassette, sub-family G (WHITE), member 4 (207593_at), score: -0.82 ABLIM3actin binding LIM protein family, member 3 (205730_s_at), score: 0.65 ADAM23ADAM metallopeptidase domain 23 (206046_at), score: 0.56 ADAMTS6ADAM metallopeptidase with thrombospondin type 1 motif, 6 (220866_at), score: -0.82 ADCY6adenylate cyclase 6 (209195_s_at), score: 0.59 AGMATagmatine ureohydrolase (agmatinase) (219792_at), score: -0.81 AIPL1aryl hydrocarbon receptor interacting protein-like 1 (219977_at), score: -0.83 ALDH1L1aldehyde dehydrogenase 1 family, member L1 (215798_at), score: -0.9 ANXA8L2annexin A8-like 2 (203074_at), score: -0.8 APOMapolipoprotein M (205682_x_at), score: -0.77 ARSAarylsulfatase A (204443_at), score: 0.59 BAT2LHLA-B associated transcript 2-like (212068_s_at), score: 0.58 BTNL3butyrophilin-like 3 (217207_s_at), score: -0.78 C1orf61chromosome 1 open reading frame 61 (205103_at), score: -0.78 C2orf54chromosome 2 open reading frame 54 (220149_at), score: -0.78 C8orf17chromosome 8 open reading frame 17 (208266_at), score: -0.78 C8orf60chromosome 8 open reading frame 60 (220712_at), score: -0.77 C9orf167chromosome 9 open reading frame 167 (219620_x_at), score: 0.67 C9orf33chromosome 9 open reading frame 33 (217130_at), score: -0.8 CA4carbonic anhydrase IV (206209_s_at), score: -0.83 CALB1calbindin 1, 28kDa (205626_s_at), score: -0.8 CAMK1calcium/calmodulin-dependent protein kinase I (204392_at), score: 0.57 CARTPTCART prepropeptide (206339_at), score: -0.79 CCBP2chemokine binding protein 2 (206887_at), score: -0.79 CEACAM1carcinoembryonic antigen-related cell adhesion molecule 1 (biliary glycoprotein) (211883_x_at), score: -0.78 CHST4carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 4 (220446_s_at), score: -0.84 CLDN1claudin 1 (218182_s_at), score: 0.55 COL11A2collagen, type XI, alpha 2 (216993_s_at), score: -0.86 CST3cystatin C (201360_at), score: 0.55 CTTNBP2NLCTTNBP2 N-terminal like (214731_at), score: -0.79 CYP2A6cytochrome P450, family 2, subfamily A, polypeptide 6 (211295_x_at), score: -0.81 DGKDdiacylglycerol kinase, delta 130kDa (208072_s_at), score: 0.56 DNAJB5DnaJ (Hsp40) homolog, subfamily B, member 5 (212817_at), score: 0.6 DNASE2deoxyribonuclease II, lysosomal (214992_s_at), score: 0.55 DSPPdentin sialophosphoprotein (221681_s_at), score: -0.78 ECE1endothelin converting enzyme 1 (201750_s_at), score: 0.64 EDARectodysplasin A receptor (220048_at), score: -0.78 EPAGearly lymphoid activation protein (217050_at), score: -0.78 ERAP2endoplasmic reticulum aminopeptidase 2 (219759_at), score: 0.58 ERLIN2ER lipid raft associated 2 (221542_s_at), score: 0.55 FAM38Bfamily with sequence similarity 38, member B (219602_s_at), score: -0.79 FAM75A3family with sequence similarity 75, member A3 (215935_at), score: -0.79 FBP1fructose-1,6-bisphosphatase 1 (209696_at), score: -0.87 FETUBfetuin B (214417_s_at), score: -0.81 FGL1fibrinogen-like 1 (205305_at), score: -0.78 FHOD1formin homology 2 domain containing 1 (218530_at), score: 0.68 GABRR1gamma-aminobutyric acid (GABA) receptor, rho 1 (206525_at), score: -0.77 GBA3glucosidase, beta, acid 3 (cytosolic) (219954_s_at), score: -0.88 GDF9growth differentiation factor 9 (221314_at), score: -0.8 GMPRguanosine monophosphate reductase (204187_at), score: 0.62 GNAO1guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O (204762_s_at), score: -0.8 GNG4guanine nucleotide binding protein (G protein), gamma 4 (205184_at), score: -0.82 GPD1Lglycerol-3-phosphate dehydrogenase 1-like (212510_at), score: 0.59 GPR32G protein-coupled receptor 32 (221469_at), score: -0.84 GPX3glutathione peroxidase 3 (plasma) (214091_s_at), score: 0.67 HAB1B1 for mucin (215778_x_at), score: -0.83 HAMPhepcidin antimicrobial peptide (220491_at), score: -0.86 HAO2hydroxyacid oxidase 2 (long chain) (220801_s_at), score: -0.77 HHEXhematopoietically expressed homeobox (215933_s_at), score: 0.67 HIST1H4Dhistone cluster 1, H4d (208076_at), score: -0.87 HLA-Amajor histocompatibility complex, class I, A (217436_x_at), score: 0.59 HLA-Cmajor histocompatibility complex, class I, C (211799_x_at), score: 0.59 HLA-Emajor histocompatibility complex, class I, E (200904_at), score: 0.72 HLA-Fmajor histocompatibility complex, class I, F (204806_x_at), score: 0.61 HOXA6homeobox A6 (208557_at), score: -0.8 HOXD11homeobox D11 (214604_at), score: -0.8 HOXD4homeobox D4 (205522_at), score: -0.78 HSF1heat shock transcription factor 1 (202344_at), score: 0.63 HTR1E5-hydroxytryptamine (serotonin) receptor 1E (207404_s_at), score: -0.77 IDUAiduronidase, alpha-L- (205059_s_at), score: 0.63 IL23Ainterleukin 23, alpha subunit p19 (211796_s_at), score: 0.55 IL9Rinterleukin 9 receptor (217212_s_at), score: -0.8 INE1inactivation escape 1 (non-protein coding) (207252_at), score: -0.77 ITKIL2-inducible T-cell kinase (211339_s_at), score: -0.77 JMJD7jumonji domain containing 7 (60528_at), score: -0.78 JUPjunction plakoglobin (201015_s_at), score: 0.56 KHDRBS3KH domain containing, RNA binding, signal transduction associated 3 (209781_s_at), score: 0.59 KLHL26kelch-like 26 (Drosophila) (219354_at), score: 0.7 KMOkynurenine 3-monooxygenase (kynurenine 3-hydroxylase) (205307_s_at), score: -0.77 LAT2linker for activation of T cells family, member 2 (211768_at), score: -0.82 LEPREL2leprecan-like 2 (204854_at), score: 0.62 LGALS14lectin, galactoside-binding, soluble, 14 (220158_at), score: -0.83 LITAFlipopolysaccharide-induced TNF factor (200706_s_at), score: 0.57 LOC149478hypothetical protein LOC149478 (215462_at), score: -0.79 LOC730227hypothetical protein LOC730227 (215756_at), score: -0.77 LPAR1lysophosphatidic acid receptor 1 (204038_s_at), score: 0.57 LRP5low density lipoprotein receptor-related protein 5 (209468_at), score: 0.55 LRRTM4leucine rich repeat transmembrane neuronal 4 (220345_at), score: -0.77 LYNv-yes-1 Yamaguchi sarcoma viral related oncogene homolog (202625_at), score: 0.56 MADCAM1mucosal vascular addressin cell adhesion molecule 1 (208037_s_at), score: -0.8 MAGOH2mago-nashi homolog 2, proliferation-associated (Drosophila) (217692_at), score: -0.8 MAN1C1mannosidase, alpha, class 1C, member 1 (218918_at), score: 0.57 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: 0.56 MBmyoglobin (204179_at), score: -0.81 MEOX1mesenchyme homeobox 1 (205619_s_at), score: -1 MTSS1metastasis suppressor 1 (203037_s_at), score: 0.56 MYH15myosin, heavy chain 15 (215331_at), score: -0.8 N4BP3Nedd4 binding protein 3 (214775_at), score: -0.81 NACAP1nascent-polypeptide-associated complex alpha polypeptide pseudogene 1 (211445_x_at), score: -0.88 NCRNA00093non-protein coding RNA 93 (210723_x_at), score: -0.78 NFKB1nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 (209239_at), score: 0.62 NID1nidogen 1 (202008_s_at), score: 0.55 NINJ1ninjurin 1 (203045_at), score: 0.67 OR7E47Polfactory receptor, family 7, subfamily E, member 47 pseudogene (216698_x_at), score: -0.81 P2RX4purinergic receptor P2X, ligand-gated ion channel, 4 (204088_at), score: 0.59 PDE5Aphosphodiesterase 5A, cGMP-specific (206757_at), score: 0.56 PDE8Bphosphodiesterase 8B (213228_at), score: -0.76 PDLIM3PDZ and LIM domain 3 (209621_s_at), score: 0.55 PECAM1platelet/endothelial cell adhesion molecule (208982_at), score: -0.87 PEG3paternally expressed 3 (209242_at), score: -0.83 PGCprogastricsin (pepsinogen C) (205261_at), score: -0.77 PHF7PHD finger protein 7 (215622_x_at), score: -0.85 PIP5K1Cphosphatidylinositol-4-phosphate 5-kinase, type I, gamma (212518_at), score: 0.61 PKNOX2PBX/knotted 1 homeobox 2 (63305_at), score: -0.79 PKP2plakophilin 2 (207717_s_at), score: 0.71 PNMAL1PNMA-like 1 (218824_at), score: 0.58 POU4F1POU class 4 homeobox 1 (206940_s_at), score: -0.93 PRB3proline-rich protein BstNI subfamily 3 (206998_x_at), score: -0.82 PROL1proline rich, lacrimal 1 (208004_at), score: -0.79 PTGDSprostaglandin D2 synthase 21kDa (brain) (212187_x_at), score: 0.61 PTPN6protein tyrosine phosphatase, non-receptor type 6 (206687_s_at), score: -0.87 RALGPS2Ral GEF with PH domain and SH3 binding motif 2 (220338_at), score: -0.82 REG3Aregenerating islet-derived 3 alpha (205815_at), score: -0.85 RETret proto-oncogene (205879_x_at), score: -0.77 RP5-886K2.1neuronal thread protein AD7c-NTP (207953_at), score: -0.81 RPS4Y1ribosomal protein S4, Y-linked 1 (201909_at), score: 0.66 RRHretinal pigment epithelium-derived rhodopsin homolog (208314_at), score: -0.82 S100A14S100 calcium binding protein A14 (218677_at), score: -0.78 SAV1salvador homolog 1 (Drosophila) (218276_s_at), score: 0.55 SCARB1scavenger receptor class B, member 1 (201819_at), score: 0.65 SCG2secretogranin II (chromogranin C) (204035_at), score: 0.63 SIGLEC1sialic acid binding Ig-like lectin 1, sialoadhesin (44673_at), score: -0.77 SIN3BSIN3 homolog B, transcription regulator (yeast) (209352_s_at), score: 0.63 SLC1A7solute carrier family 1 (glutamate transporter), member 7 (210923_at), score: -0.82 SLC26A10solute carrier family 26, member 10 (214951_at), score: -0.83 SLC4A5solute carrier family 4, sodium bicarbonate cotransporter, member 5 (204296_at), score: -0.85 SLC9A3R2solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 (209830_s_at), score: 0.7 SPARCL1SPARC-like 1 (hevin) (200795_at), score: -0.84 STOMstomatin (201060_x_at), score: 0.63 SULT1B1sulfotransferase family, cytosolic, 1B, member 1 (207601_at), score: -0.85 SYT1synaptotagmin I (203999_at), score: 0.55 TAS2R16taste receptor, type 2, member 16 (221444_at), score: -0.8 TERF2telomeric repeat binding factor 2 (203611_at), score: 0.59 TGM4transglutaminase 4 (prostate) (217566_s_at), score: -0.84 TM6SF1transmembrane 6 superfamily member 1 (219892_at), score: 0.63 TNFAIP2tumor necrosis factor, alpha-induced protein 2 (202510_s_at), score: 0.56 TNFSF4tumor necrosis factor (ligand) superfamily, member 4 (207426_s_at), score: 0.6 TNK1tyrosine kinase, non-receptor, 1 (217149_x_at), score: -0.77 TPD52tumor protein D52 (201690_s_at), score: 0.63 TPP1tripeptidyl peptidase I (200742_s_at), score: 0.55 TRA@T cell receptor alpha locus (216540_at), score: -0.82 TTTY9Atestis-specific transcript, Y-linked 9A (211460_at), score: -0.77 VEGFBvascular endothelial growth factor B (203683_s_at), score: 0.57 WNT6wingless-type MMTV integration site family, member 6 (71933_at), score: -0.77 ZKSCAN3zinc finger with KRAB and SCAN domains 3 (211773_s_at), score: -0.8 ZNF767zinc finger family member 767 (219627_at), score: -0.79 ZPBPzona pellucida binding protein (207021_at), score: -0.88
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| 2Twin.CEL | 2 | 2 | DS-twin | hgu133plus2 | none | DS-twin 2 |
| t21d 08-03.CEL | 7 | 1 | DS-CC | hgu133a | Down | DS-CC 7 |
| 6Twin.CEL | 6 | 2 | DS-twin | hgu133plus2 | none | DS-twin 6 |
| E-GEOD-3407-raw-cel-1437949557.cel | 1 | 4 | Cockayne | hgu133a | CS | eGFP |
| t21a 08-03.CEL | 4 | 1 | DS-CC | hgu133a | Down | DS-CC 4 |
| 4Twin.CEL | 4 | 2 | DS-twin | hgu133plus2 | none | DS-twin 4 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 46A.CEL | 1 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |