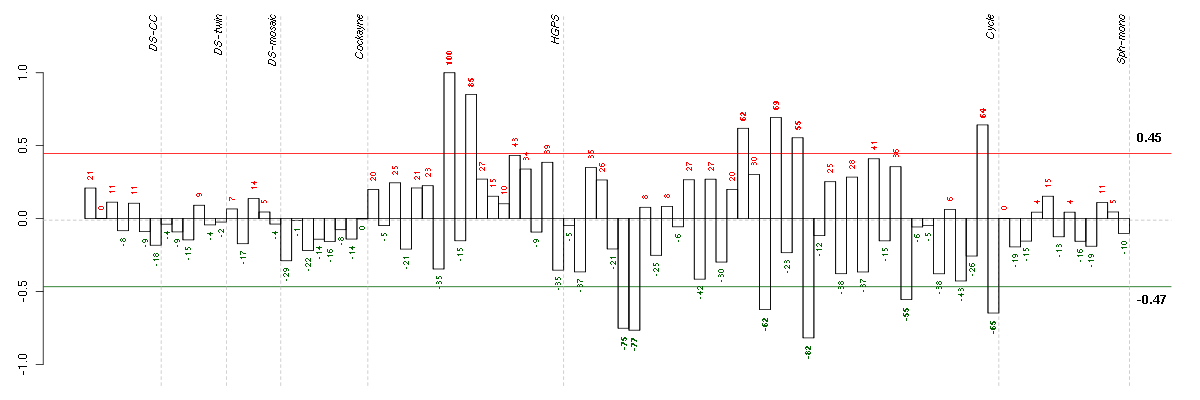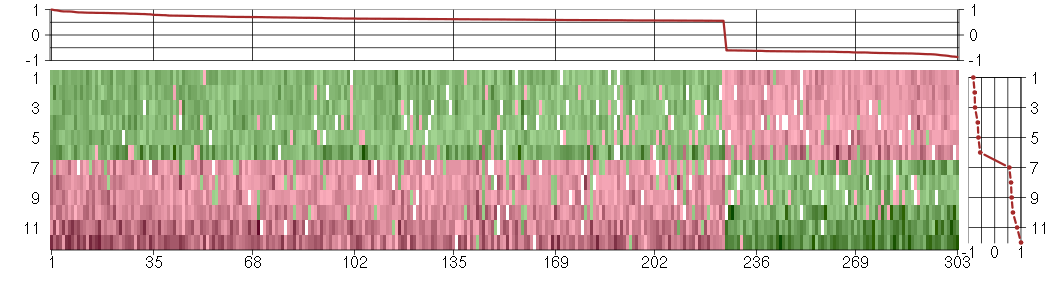

Under-expression is coded with green,
over-expression with red color.



ADCK2aarF domain containing kinase 2 (221893_s_at), score: 0.86 ADCY3adenylate cyclase 3 (209321_s_at), score: 0.63 ARFGAP1ADP-ribosylation factor GTPase activating protein 1 (217888_s_at), score: 0.6 ARFGAP2ADP-ribosylation factor GTPase activating protein 2 (211975_at), score: 0.58 ARHGAP22Rho GTPase activating protein 22 (206298_at), score: 0.66 ARL17P1ADP-ribosylation factor-like 17 pseudogene 1 (210718_s_at), score: 0.59 ARMCX1armadillo repeat containing, X-linked 1 (218694_at), score: -0.64 ARPC5Lactin related protein 2/3 complex, subunit 5-like (220966_x_at), score: 0.61 ATF2activating transcription factor 2 (212984_at), score: -0.63 ATN1atrophin 1 (40489_at), score: 0.88 ATXN1ataxin 1 (203232_s_at), score: -0.8 B3GALT2UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 (217452_s_at), score: -0.66 B3GAT3beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) (203452_at), score: 0.81 B4GALT5UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 (221484_at), score: 0.65 BACE2beta-site APP-cleaving enzyme 2 (217867_x_at), score: 0.78 BAIAP2BAI1-associated protein 2 (205294_at), score: 0.85 BAT2HLA-B associated transcript 2 (212081_x_at), score: 0.59 BCAT1branched chain aminotransferase 1, cytosolic (214452_at), score: -0.65 BCL7BB-cell CLL/lymphoma 7B (202518_at), score: 0.64 BIN1bridging integrator 1 (210201_x_at), score: 0.56 BLCAPbladder cancer associated protein (201032_at), score: 0.6 BRD9bromodomain containing 9 (220155_s_at), score: 0.78 BRMS1breast cancer metastasis suppressor 1 (215631_s_at), score: 0.58 BST1bone marrow stromal cell antigen 1 (205715_at), score: -0.6 BTBD2BTB (POZ) domain containing 2 (207722_s_at), score: 0.67 BTN3A3butyrophilin, subfamily 3, member A3 (204820_s_at), score: -0.67 C14orf104chromosome 14 open reading frame 104 (219166_at), score: -0.62 C16orf57chromosome 16 open reading frame 57 (218060_s_at), score: 0.96 C20orf111chromosome 20 open reading frame 111 (209020_at), score: -0.65 C20orf30chromosome 20 open reading frame 30 (220477_s_at), score: -0.64 C22orf29chromosome 22 open reading frame 29 (219560_at), score: 0.57 C2CD2LC2CD2-like (204757_s_at), score: 0.61 C2orf43chromosome 2 open reading frame 43 (222192_s_at), score: -0.66 C5orf28chromosome 5 open reading frame 28 (219029_at), score: -0.67 CABIN1calcineurin binding protein 1 (37652_at), score: 0.56 CASKcalcium/calmodulin-dependent serine protein kinase (MAGUK family) (211208_s_at), score: 0.59 CASKIN2CASK interacting protein 2 (61297_at), score: 0.59 CCND3cyclin D3 (201700_at), score: 0.62 CCNE1cyclin E1 (213523_at), score: 0.76 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (217849_s_at), score: 0.81 CDCA4cell division cycle associated 4 (218399_s_at), score: 0.56 CDKN1Bcyclin-dependent kinase inhibitor 1B (p27, Kip1) (209112_at), score: -0.69 CDR2Lcerebellar degeneration-related protein 2-like (213230_at), score: 0.86 CENPTcentromere protein T (218148_at), score: 0.84 CEP164centrosomal protein 164kDa (204251_s_at), score: 0.67 CHST3carbohydrate (chondroitin 6) sulfotransferase 3 (209834_at), score: 0.76 CICcapicua homolog (Drosophila) (212784_at), score: 0.69 CITcitron (rho-interacting, serine/threonine kinase 21) (212801_at), score: 0.66 CIZ1CDKN1A interacting zinc finger protein 1 (205516_x_at), score: 0.68 CLCF1cardiotrophin-like cytokine factor 1 (219500_at), score: 0.69 CLIC4chloride intracellular channel 4 (201559_s_at), score: -0.71 CLIP1CAP-GLY domain containing linker protein 1 (210716_s_at), score: -0.65 CLIP2CAP-GLY domain containing linker protein 2 (211031_s_at), score: 0.71 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: 0.75 COL10A1collagen, type X, alpha 1 (217428_s_at), score: 0.61 COL7A1collagen, type VII, alpha 1 (217312_s_at), score: 0.87 COL8A1collagen, type VIII, alpha 1 (214587_at), score: -0.64 COPS7ACOP9 constitutive photomorphogenic homolog subunit 7A (Arabidopsis) (209029_at), score: 0.57 CORINcorin, serine peptidase (220356_at), score: -0.67 CORO2Bcoronin, actin binding protein, 2B (209789_at), score: 0.69 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: 0.62 CYTH1cytohesin 1 (202880_s_at), score: 0.71 DCAKDdephospho-CoA kinase domain containing (221224_s_at), score: -0.61 DCHS1dachsous 1 (Drosophila) (222101_s_at), score: 0.59 DDX3XDEAD (Asp-Glu-Ala-Asp) box polypeptide 3, X-linked (201211_s_at), score: -0.61 DDX54DEAD (Asp-Glu-Ala-Asp) box polypeptide 54 (219111_s_at), score: 0.82 DENND4BDENN/MADD domain containing 4B (202860_at), score: 0.63 DMWDdystrophia myotonica, WD repeat containing (33768_at), score: 0.75 DNM1dynamin 1 (215116_s_at), score: 0.61 DNM2dynamin 2 (202253_s_at), score: 0.59 DOK4docking protein 4 (209691_s_at), score: 0.73 EDIL3EGF-like repeats and discoidin I-like domains 3 (207379_at), score: -0.77 EIF2S3eukaryotic translation initiation factor 2, subunit 3 gamma, 52kDa (205321_at), score: -0.84 EMP2epithelial membrane protein 2 (204975_at), score: 0.58 ENO2enolase 2 (gamma, neuronal) (201313_at), score: 0.58 ENO3enolase 3 (beta, muscle) (204483_at), score: -0.72 EPS15epidermal growth factor receptor pathway substrate 15 (217887_s_at), score: -0.72 F11coagulation factor XI (206610_s_at), score: -0.65 F2Rcoagulation factor II (thrombin) receptor (203989_x_at), score: -0.7 F3coagulation factor III (thromboplastin, tissue factor) (204363_at), score: 0.57 FAM168Bfamily with sequence similarity 168, member B (212017_at), score: 0.86 FAM38Afamily with sequence similarity 38, member A (202771_at), score: 0.6 FAM3Afamily with sequence similarity 3, member A (38043_at), score: 0.61 FASNfatty acid synthase (212218_s_at), score: 0.63 FCF1FCF1 small subunit (SSU) processome component homolog (S. cerevisiae) (219927_at), score: -0.7 FHfumarate hydratase (203032_s_at), score: -0.64 FHL1four and a half LIM domains 1 (201539_s_at), score: -0.72 FKRPfukutin related protein (219853_at), score: 0.65 FKSG2apoptosis inhibitor (208588_at), score: -0.62 FLJ12529pre-mRNA cleavage factor I, 59 kDa subunit (217866_at), score: 0.97 FNDC4fibronectin type III domain containing 4 (218843_at), score: 0.57 FOSL1FOS-like antigen 1 (204420_at), score: 0.64 FOXE1forkhead box E1 (thyroid transcription factor 2) (206912_at), score: -0.68 FOXK2forkhead box K2 (203064_s_at), score: 0.83 FURINfurin (paired basic amino acid cleaving enzyme) (201945_at), score: 0.87 FZR1fizzy/cell division cycle 20 related 1 (Drosophila) (209416_s_at), score: 0.59 GAAglucosidase, alpha; acid (202812_at), score: 0.6 GAKcyclin G associated kinase (202281_at), score: 0.66 GALNT6UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 6 (GalNAc-T6) (219956_at), score: 0.6 GGNBP2gametogenetin binding protein 2 (218079_s_at), score: 0.65 GIT1G protein-coupled receptor kinase interacting ArfGAP 1 (218030_at), score: 0.76 GOLGA3golgi autoantigen, golgin subfamily a, 3 (202106_at), score: 0.62 GP1BBglycoprotein Ib (platelet), beta polypeptide (206655_s_at), score: 0.6 GPR172AG protein-coupled receptor 172A (222155_s_at), score: 0.57 GRINAglutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) (212090_at), score: 0.65 GTF2Igeneral transcription factor II, i (210892_s_at), score: -0.71 GYS1glycogen synthase 1 (muscle) (201673_s_at), score: 0.58 HBP1HMG-box transcription factor 1 (209102_s_at), score: -0.65 HDAC7histone deacetylase 7 (217937_s_at), score: 0.67 HECTD3HECT domain containing 3 (218632_at), score: 0.57 HNRNPA3P1heterogeneous nuclear ribonucleoprotein A3 pseudogene 1 (206809_s_at), score: 0.57 HNRNPH2heterogeneous nuclear ribonucleoprotein H2 (H') (201132_at), score: -0.86 HNRNPUL1heterogeneous nuclear ribonucleoprotein U-like 1 (209675_s_at), score: 0.57 HOXC11homeobox C11 (206745_at), score: 0.73 HPS6Hermansky-Pudlak syndrome 6 (219052_at), score: 0.57 HSPA12Aheat shock 70kDa protein 12A (214434_at), score: 0.61 IDUAiduronidase, alpha-L- (205059_s_at), score: 0.83 IFI16interferon, gamma-inducible protein 16 (208965_s_at), score: -0.66 IFNGR1interferon gamma receptor 1 (211676_s_at), score: -0.67 IL17RAinterleukin 17 receptor A (205707_at), score: 0.67 IPO8importin 8 (205701_at), score: -0.62 IPPKinositol 1,3,4,5,6-pentakisphosphate 2-kinase (219092_s_at), score: 0.57 ITGA5integrin, alpha 5 (fibronectin receptor, alpha polypeptide) (201389_at), score: 0.57 JAG1jagged 1 (Alagille syndrome) (216268_s_at), score: -0.69 KCNE4potassium voltage-gated channel, Isk-related family, member 4 (222379_at), score: -0.68 KCNJ15potassium inwardly-rectifying channel, subfamily J, member 15 (210119_at), score: 0.58 KCNN2potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2 (220116_at), score: -0.61 KCNN4potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 (204401_at), score: 0.6 KCTD13potassium channel tetramerisation domain containing 13 (45653_at), score: 0.63 KCTD2potassium channel tetramerisation domain containing 2 (212564_at), score: 0.56 KITv-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog (205051_s_at), score: -0.65 KLC2kinesin light chain 2 (218906_x_at), score: 0.6 KLF9Kruppel-like factor 9 (203543_s_at), score: -0.7 KPNA6karyopherin alpha 6 (importin alpha 7) (212101_at), score: 0.56 LEPREL2leprecan-like 2 (204854_at), score: 0.62 LIG1ligase I, DNA, ATP-dependent (202726_at), score: 0.56 LMF2lipase maturation factor 2 (212682_s_at), score: 0.72 LOC100133105hypothetical protein LOC100133105 (214237_x_at), score: -0.74 LOC100133166similar to neighbor of BRCA1 gene 1 (201383_s_at), score: -0.62 LOC339047hypothetical protein LOC339047 (221501_x_at), score: 0.61 LOC90379hypothetical protein BC002926 (221849_s_at), score: 0.79 LRDDleucine-rich repeats and death domain containing (221640_s_at), score: 0.63 LRFN4leucine rich repeat and fibronectin type III domain containing 4 (219491_at), score: 0.74 LRP1low density lipoprotein-related protein 1 (alpha-2-macroglobulin receptor) (200785_s_at), score: 0.57 LRRC37A2leucine rich repeat containing 37, member A2 (221740_x_at), score: -0.75 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: 1 MAP2K3mitogen-activated protein kinase kinase 3 (207667_s_at), score: 0.57 MAP3K6mitogen-activated protein kinase kinase kinase 6 (219278_at), score: 0.68 MAP7D1MAP7 domain containing 1 (217943_s_at), score: 0.83 MAPK7mitogen-activated protein kinase 7 (35617_at), score: 0.56 MAPKAPK3mitogen-activated protein kinase-activated protein kinase 3 (202788_at), score: 0.68 MAST2microtubule associated serine/threonine kinase 2 (211593_s_at), score: 0.87 MAST4microtubule associated serine/threonine kinase family member 4 (40016_g_at), score: 0.61 MAXMYC associated factor X (210734_x_at), score: -0.6 MBD3methyl-CpG binding domain protein 3 (41160_at), score: 0.62 MBNL2muscleblind-like 2 (Drosophila) (205018_s_at), score: -0.85 MCM3APminichromosome maintenance complex component 3 associated protein (212269_s_at), score: 0.57 MED15mediator complex subunit 15 (222175_s_at), score: 0.68 MGAT1mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase (201126_s_at), score: 0.61 MICAL3microtubule associated monoxygenase, calponin and LIM domain containing 3 (212715_s_at), score: 0.79 MINK1misshapen-like kinase 1 (zebrafish) (214246_x_at), score: 0.76 MLL4myeloid/lymphoid or mixed-lineage leukemia 4 (203419_at), score: 0.71 MR1major histocompatibility complex, class I-related (207565_s_at), score: -0.73 MSL1male-specific lethal 1 homolog (Drosophila) (212708_at), score: 0.63 MTA2metastasis associated 1 family, member 2 (203442_x_at), score: 0.58 MUL1mitochondrial E3 ubiquitin ligase 1 (218246_at), score: 0.7 MYO1Bmyosin IB (212364_at), score: -0.64 MYO9Bmyosin IXB (217297_s_at), score: 0.65 MYST3MYST histone acetyltransferase (monocytic leukemia) 3 (202423_at), score: 0.69 NBL1neuroblastoma, suppression of tumorigenicity 1 (37005_at), score: 0.92 NCLNnicalin homolog (zebrafish) (222206_s_at), score: 0.58 NCOR2nuclear receptor co-repressor 2 (207760_s_at), score: 0.64 NDUFA5NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5, 13kDa (201304_at), score: -0.66 NET1neuroepithelial cell transforming 1 (201830_s_at), score: 0.56 NFATC2IPnuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 interacting protein (217526_at), score: 0.61 NFATC4nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 (205897_at), score: 0.75 NFYBnuclear transcription factor Y, beta (218129_s_at), score: -0.61 NKX3-1NK3 homeobox 1 (209706_at), score: 0.87 NPHP4nephronophthisis 4 (213471_at), score: 0.63 NPIPnuclear pore complex interacting protein (204538_x_at), score: 0.56 NRG1neuregulin 1 (206343_s_at), score: 0.57 OLFM1olfactomedin 1 (213131_at), score: 0.68 OR7E156Polfactory receptor, family 7, subfamily E, member 156 pseudogene (222327_x_at), score: -0.65 ORAI2ORAI calcium release-activated calcium modulator 2 (218812_s_at), score: 0.62 OTUD3OTU domain containing 3 (213216_at), score: 0.63 PAFAH1B2platelet-activating factor acetylhydrolase, isoform Ib, beta subunit 30kDa (210160_at), score: -0.73 PARVBparvin, beta (37965_at), score: 0.68 PCCApropionyl Coenzyme A carboxylase, alpha polypeptide (203860_at), score: -0.64 PCDHG@protocadherin gamma cluster (215836_s_at), score: 0.7 PCDHGA1protocadherin gamma subfamily A, 1 (209079_x_at), score: 0.87 PCGF2polycomb group ring finger 2 (203793_x_at), score: 0.58 PDCD4programmed cell death 4 (neoplastic transformation inhibitor) (202731_at), score: -0.72 PDIA3protein disulfide isomerase family A, member 3 (208612_at), score: -0.61 PDLIM4PDZ and LIM domain 4 (211564_s_at), score: 0.7 PEX26peroxisomal biogenesis factor 26 (219180_s_at), score: 0.63 PIP5K1Cphosphatidylinositol-4-phosphate 5-kinase, type I, gamma (212518_at), score: 0.92 PKMYT1protein kinase, membrane associated tyrosine/threonine 1 (204267_x_at), score: 0.65 PKN1protein kinase N1 (202161_at), score: 0.63 PLAURplasminogen activator, urokinase receptor (211924_s_at), score: 0.7 PLCD1phospholipase C, delta 1 (205125_at), score: 0.64 PLCXD1phosphatidylinositol-specific phospholipase C, X domain containing 1 (218951_s_at), score: 0.78 PLEKHJ1pleckstrin homology domain containing, family J member 1 (218290_at), score: 0.56 PLXNA3plexin A3 (203623_at), score: 0.59 PNPLA6patatin-like phospholipase domain containing 6 (203718_at), score: 0.58 POLEpolymerase (DNA directed), epsilon (216026_s_at), score: 0.63 POLIpolymerase (DNA directed) iota (219317_at), score: -0.72 POLR2Apolymerase (RNA) II (DNA directed) polypeptide A, 220kDa (202725_at), score: 0.71 POLR2Dpolymerase (RNA) II (DNA directed) polypeptide D (203664_s_at), score: 0.6 POLSpolymerase (DNA directed) sigma (202466_at), score: 0.94 POM121POM121 membrane glycoprotein (rat) (212178_s_at), score: 0.9 POM121CPOM121 membrane glycoprotein C (213360_s_at), score: 0.65 POMZP3POM (POM121 homolog, rat) and ZP3 fusion (204148_s_at), score: 0.62 PQLC1PQ loop repeat containing 1 (218208_at), score: 0.58 PRKACAprotein kinase, cAMP-dependent, catalytic, alpha (202801_at), score: 0.63 PRKCDprotein kinase C, delta (202545_at), score: 0.61 PRR14proline rich 14 (218714_at), score: 0.58 PTGS1prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) (215813_s_at), score: 0.58 PTPN9protein tyrosine phosphatase, non-receptor type 9 (202958_at), score: 0.74 RAB11FIP3RAB11 family interacting protein 3 (class II) (203933_at), score: 0.64 RAB11FIP5RAB11 family interacting protein 5 (class I) (210879_s_at), score: 0.71 RAB15RAB15, member RAS onocogene family (221810_at), score: 0.82 RAB22ARAB22A, member RAS oncogene family (218360_at), score: 0.62 RAB35RAB35, member RAS oncogene family (205461_at), score: -0.65 RAB36RAB36, member RAS oncogene family (211471_s_at), score: 0.61 RAB5ARAB5A, member RAS oncogene family (206113_s_at), score: -0.72 RAD54L2RAD54-like 2 (S. cerevisiae) (213205_s_at), score: 0.87 RBM15BRNA binding motif protein 15B (202689_at), score: 0.68 RDXradixin (204969_s_at), score: -0.65 RHOBTB1Rho-related BTB domain containing 1 (212651_at), score: -0.71 RNF126ring finger protein 126 (205748_s_at), score: 0.56 RNF19Bring finger protein 19B (213038_at), score: 0.61 RNF220ring finger protein 220 (219988_s_at), score: 0.76 RPL10Lribosomal protein L10-like (217559_at), score: -0.64 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: -0.81 RPRD2regulation of nuclear pre-mRNA domain containing 2 (212553_at), score: 0.58 RPS17P5ribosomal protein S17 pseudogene 5 (216348_at), score: -0.76 RSL24D1ribosomal L24 domain containing 1 (217915_s_at), score: -0.79 SCAMP1secretory carrier membrane protein 1 (206667_s_at), score: -0.75 SCAMP4secretory carrier membrane protein 4 (213244_at), score: 0.88 SCN9Asodium channel, voltage-gated, type IX, alpha subunit (206950_at), score: -0.65 SCRN3secernin 3 (219234_x_at), score: -0.62 SDC4syndecan 4 (202071_at), score: 0.57 SETBP1SET binding protein 1 (205933_at), score: 0.58 SF1splicing factor 1 (208313_s_at), score: 0.85 SF3A2splicing factor 3a, subunit 2, 66kDa (37462_i_at), score: 0.69 SIPA1L3signal-induced proliferation-associated 1 like 3 (213600_at), score: 0.73 SKIV2Lsuperkiller viralicidic activity 2-like (S. cerevisiae) (203727_at), score: 0.56 SLC25A22solute carrier family 25 (mitochondrial carrier: glutamate), member 22 (218725_at), score: 0.56 SLC6A10Psolute carrier family 6 (neurotransmitter transporter, creatine), member 10 (pseudogene) (215812_s_at), score: 0.62 SLC6A8solute carrier family 6 (neurotransmitter transporter, creatine), member 8 (202219_at), score: 0.7 SMARCD1SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 (209518_at), score: 0.57 SMG7Smg-7 homolog, nonsense mediated mRNA decay factor (C. elegans) (201794_s_at), score: 0.71 SMYD5SMYD family member 5 (209516_at), score: 0.64 SNAP23synaptosomal-associated protein, 23kDa (214544_s_at), score: -0.82 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: 0.92 SPG7spastic paraplegia 7 (pure and complicated autosomal recessive) (202104_s_at), score: 0.6 SPHK1sphingosine kinase 1 (219257_s_at), score: 0.62 SPON1spondin 1, extracellular matrix protein (209436_at), score: 0.7 SPRY4sprouty homolog 4 (Drosophila) (221489_s_at), score: 0.72 SSBP3single stranded DNA binding protein 3 (217991_x_at), score: 0.63 STARD8StAR-related lipid transfer (START) domain containing 8 (206868_at), score: 0.61 STRN4striatin, calmodulin binding protein 4 (217903_at), score: 0.92 SUPT6Hsuppressor of Ty 6 homolog (S. cerevisiae) (208831_x_at), score: 0.64 TAF9BTAF9B RNA polymerase II, TATA box binding protein (TBP)-associated factor, 31kDa (221618_s_at), score: -0.64 TAOK2TAO kinase 2 (204986_s_at), score: 0.64 TAPBPTAP binding protein (tapasin) (208829_at), score: 0.74 TATDN2TatD DNase domain containing 2 (203648_at), score: 0.63 TBC1D10BTBC1 domain family, member 10B (220947_s_at), score: 0.75 TBCDtubulin folding cofactor D (211052_s_at), score: 0.64 TBPTATA box binding protein (203135_at), score: 0.62 TBX3T-box 3 (219682_s_at), score: 0.64 TCF7transcription factor 7 (T-cell specific, HMG-box) (205255_x_at), score: 0.73 TELO2TEL2, telomere maintenance 2, homolog (S. cerevisiae) (34260_at), score: 0.61 TGFAtransforming growth factor, alpha (205016_at), score: 0.69 THUMPD2THUMP domain containing 2 (219248_at), score: 0.64 TMED7transmembrane emp24 protein transport domain containing 7 (209404_s_at), score: -0.61 TMEM123transmembrane protein 123 (211967_at), score: -0.75 TMEM158transmembrane protein 158 (213338_at), score: 0.71 TMEM22transmembrane protein 22 (219569_s_at), score: 0.65 TNK2tyrosine kinase, non-receptor, 2 (203839_s_at), score: 0.67 TOXthymocyte selection-associated high mobility group box (204529_s_at), score: -0.71 TSKUtsukushin (218245_at), score: 0.6 TXLNAtaxilin alpha (212300_at), score: 0.83 TYRO3TYRO3 protein tyrosine kinase (211432_s_at), score: 0.59 UBE2Oubiquitin-conjugating enzyme E2O (218141_at), score: 0.57 UBE2Zubiquitin-conjugating enzyme E2Z (217750_s_at), score: 0.62 UBE3Aubiquitin protein ligase E3A (213291_s_at), score: 0.59 UPF1UPF1 regulator of nonsense transcripts homolog (yeast) (211168_s_at), score: 0.57 UTRNutrophin (213022_s_at), score: -0.68 VEGFBvascular endothelial growth factor B (203683_s_at), score: 0.71 VPS37Bvacuolar protein sorting 37 homolog B (S. cerevisiae) (221704_s_at), score: 0.85 WIZwidely interspaced zinc finger motifs (52005_at), score: 0.63 WNT2Bwingless-type MMTV integration site family, member 2B (206459_s_at), score: -0.63 XAB2XPA binding protein 2 (218110_at), score: 0.73 ZDHHC18zinc finger, DHHC-type containing 18 (212860_at), score: 0.85 ZFP36L1zinc finger protein 36, C3H type-like 1 (211965_at), score: 0.56 ZFYVE26zinc finger, FYVE domain containing 26 (37943_at), score: -0.71 ZNF277zinc finger protein 277 (218645_at), score: -0.74 ZNF280Dzinc finger protein 280D (221213_s_at), score: -0.68 ZNF282zinc finger protein 282 (212892_at), score: 0.58 ZNF318zinc finger protein 318 (203521_s_at), score: 0.74 ZNF33Bzinc finger protein 33B (215022_x_at), score: -0.64 ZNF536zinc finger protein 536 (206403_at), score: 0.89
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486011.cel | 19 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486271.cel | 32 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486071.cel | 22 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485971.cel | 17 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486411.cel | 39 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486031.cel | 20 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690344.cel | 10 | 5 | HGPS | hgu133a | none | GM00038C |
| E-GEOD-3860-raw-cel-1561690304.cel | 8 | 5 | HGPS | hgu133a | none | GMO8398C |