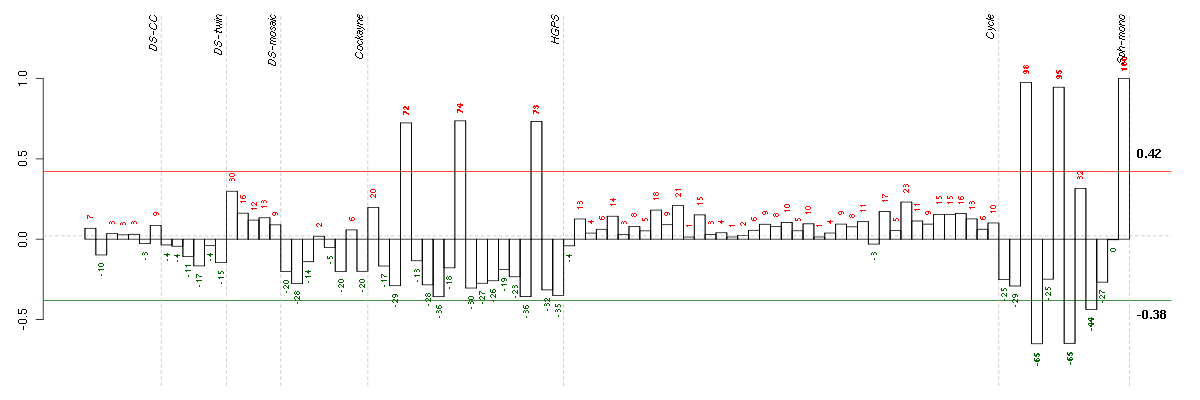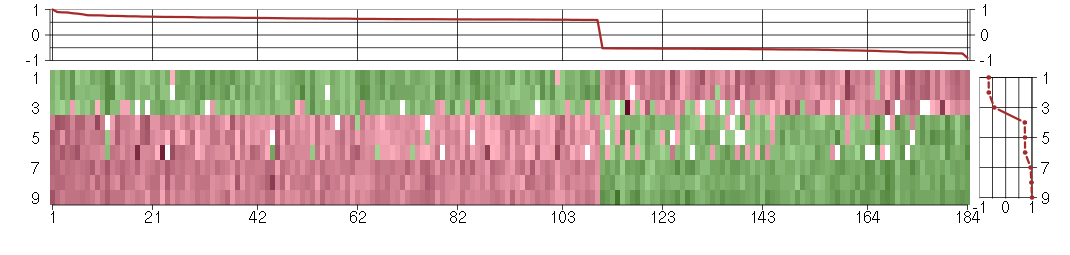

Under-expression is coded with green,
over-expression with red color.


plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
integral to membrane
Penetrating at least one phospholipid bilayer of a membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer. When used to describe a protein, indicates that all or part of the peptide sequence is embedded in the membrane.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.

ABHD2abhydrolase domain containing 2 (221815_at), score: -0.56 ACTG2actin, gamma 2, smooth muscle, enteric (202274_at), score: 0.64 ADAM19ADAM metallopeptidase domain 19 (meltrin beta) (209765_at), score: 0.61 AEBP1AE binding protein 1 (201792_at), score: 0.64 AGTR1angiotensin II receptor, type 1 (205357_s_at), score: 0.6 ALDH1A3aldehyde dehydrogenase 1 family, member A3 (203180_at), score: -0.53 AOX1aldehyde oxidase 1 (205083_at), score: 0.61 APH1Banterior pharynx defective 1 homolog B (C. elegans) (221036_s_at), score: 0.62 ARID5BAT rich interactive domain 5B (MRF1-like) (212614_at), score: 0.61 ARL15ADP-ribosylation factor-like 15 (219842_at), score: 0.68 ARSJarylsulfatase family, member J (219973_at), score: 0.62 ATP2B4ATPase, Ca++ transporting, plasma membrane 4 (212135_s_at), score: 0.69 AXLAXL receptor tyrosine kinase (202685_s_at), score: 0.62 B4GALT5UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 (221484_at), score: -0.53 BGNbiglycan (213905_x_at), score: 0.65 BTN3A1butyrophilin, subfamily 3, member A1 (209770_at), score: 0.65 C19orf2chromosome 19 open reading frame 2 (214173_x_at), score: -0.57 C21orf7chromosome 21 open reading frame 7 (221211_s_at), score: 0.7 C7orf64chromosome 7 open reading frame 64 (221596_s_at), score: -0.61 C9orf95chromosome 9 open reading frame 95 (219147_s_at), score: 0.65 CABC1chaperone, ABC1 activity of bc1 complex homolog (S. pombe) (218168_s_at), score: -0.68 CAPNS1calpain, small subunit 1 (200001_at), score: 0.6 CBFA2T2core-binding factor, runt domain, alpha subunit 2; translocated to, 2 (207625_s_at), score: -0.56 CCDC92coiled-coil domain containing 92 (218175_at), score: 0.7 CD44CD44 molecule (Indian blood group) (210916_s_at), score: -0.69 CD55CD55 molecule, decay accelerating factor for complement (Cromer blood group) (201925_s_at), score: -0.71 CDKN1Ccyclin-dependent kinase inhibitor 1C (p57, Kip2) (213183_s_at), score: -0.53 CELSR3cadherin, EGF LAG seven-pass G-type receptor 3 (flamingo homolog, Drosophila) (40020_at), score: -0.62 CIRCBF1 interacting corepressor (209571_at), score: -0.53 CLEC2BC-type lectin domain family 2, member B (209732_at), score: -0.9 CNTNAP1contactin associated protein 1 (219400_at), score: 0.68 COBLL1COBL-like 1 (203642_s_at), score: 0.78 COL4A5collagen, type IV, alpha 5 (213110_s_at), score: 0.64 CORINcorin, serine peptidase (220356_at), score: 0.71 CREB3L1cAMP responsive element binding protein 3-like 1 (213059_at), score: 0.6 CROTcarnitine O-octanoyltransferase (204573_at), score: -0.53 CRYABcrystallin, alpha B (209283_at), score: 0.62 CSRP2cysteine and glycine-rich protein 2 (211126_s_at), score: 0.63 CTGFconnective tissue growth factor (209101_at), score: 0.61 CYB561cytochrome b-561 (209163_at), score: -0.57 CYFIP2cytoplasmic FMR1 interacting protein 2 (215785_s_at), score: 0.6 DAAM2dishevelled associated activator of morphogenesis 2 (212793_at), score: 0.72 DDAH1dimethylarginine dimethylaminohydrolase 1 (209094_at), score: 0.65 DENND5BDENN/MADD domain containing 5B (215058_at), score: -0.55 DGCR2DiGeorge syndrome critical region gene 2 (214198_s_at), score: -0.55 DGKDdiacylglycerol kinase, delta 130kDa (208072_s_at), score: -0.62 DIDO1death inducer-obliterator 1 (218325_s_at), score: -0.56 EFHD1EF-hand domain family, member D1 (209343_at), score: 0.84 EHD3EH-domain containing 3 (218935_at), score: 0.67 EIF1eukaryotic translation initiation factor 1 (212130_x_at), score: -0.54 EPB41L1erythrocyte membrane protein band 4.1-like 1 (212339_at), score: -0.72 EPHB6EPH receptor B6 (204718_at), score: 0.61 EREGepiregulin (205767_at), score: 0.65 EXT1exostoses (multiple) 1 (201995_at), score: 0.75 FAM117Afamily with sequence similarity 117, member A (221249_s_at), score: -0.58 FAM174Bfamily with sequence similarity 174, member B (51158_at), score: 0.66 FIG4FIG4 homolog (S. cerevisiae) (203656_at), score: 0.62 FLNAfilamin A, alpha (actin binding protein 280) (213746_s_at), score: 0.59 FLT1fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor) (222033_s_at), score: 0.62 FOXE1forkhead box E1 (thyroid transcription factor 2) (206912_at), score: 0.75 FOXL2forkhead box L2 (220102_at), score: -0.55 FRYfurry homolog (Drosophila) (204072_s_at), score: 0.62 GAL3ST4galactose-3-O-sulfotransferase 4 (219815_at), score: 0.62 GALNT3UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3) (203397_s_at), score: 0.9 GFRA1GDNF family receptor alpha 1 (205696_s_at), score: 0.62 GLRBglycine receptor, beta (205280_at), score: 0.59 GPRC5BG protein-coupled receptor, family C, group 5, member B (203632_s_at), score: -0.72 GREM1gremlin 1, cysteine knot superfamily, homolog (Xenopus laevis) (218469_at), score: 0.75 GRIK2glutamate receptor, ionotropic, kainate 2 (213845_at), score: 0.67 GTDC1glycosyltransferase-like domain containing 1 (219770_at), score: 0.73 HERC2P2hect domain and RLD 2 pseudogene 2 (217317_s_at), score: -0.53 HEY1hairy/enhancer-of-split related with YRPW motif 1 (44783_s_at), score: -0.65 HLA-DRB1major histocompatibility complex, class II, DR beta 1 (209312_x_at), score: -0.53 HTATIP2HIV-1 Tat interactive protein 2, 30kDa (209448_at), score: 0.64 IL23Ainterleukin 23, alpha subunit p19 (211796_s_at), score: 0.71 ILKintegrin-linked kinase (201234_at), score: 0.59 IMAASLC7A5 pseudogene (208118_x_at), score: -0.69 INAinternexin neuronal intermediate filament protein, alpha (204465_s_at), score: 0.77 IQCKIQ motif containing K (213392_at), score: -0.7 ISL1ISL LIM homeobox 1 (206104_at), score: 0.62 KANK2KN motif and ankyrin repeat domains 2 (218418_s_at), score: 0.61 KCNK1potassium channel, subfamily K, member 1 (204679_at), score: -0.54 KCTD2potassium channel tetramerisation domain containing 2 (212564_at), score: -0.54 KYNUkynureninase (L-kynurenine hydrolase) (217388_s_at), score: -0.64 LDLRlow density lipoprotein receptor (202068_s_at), score: 0.59 LEF1lymphoid enhancer-binding factor 1 (221558_s_at), score: -0.7 LIN7Alin-7 homolog A (C. elegans) (206440_at), score: 0.61 LMO7LIM domain 7 (202674_s_at), score: 0.61 LOC440354PI-3-kinase-related kinase SMG-1 pseudogene (210396_s_at), score: -0.52 LPPR4plasticity related gene 1 (213496_at), score: 0.88 LRP4low density lipoprotein receptor-related protein 4 (212850_s_at), score: -0.61 LRRC16Aleucine rich repeat containing 16A (219573_at), score: 0.71 LUMlumican (201744_s_at), score: 0.71 MAB21L1mab-21-like 1 (C. elegans) (206163_at), score: 0.65 MAFBv-maf musculoaponeurotic fibrosarcoma oncogene homolog B (avian) (218559_s_at), score: -0.68 MAMLD1mastermind-like domain containing 1 (205088_at), score: 0.63 MANSC1MANSC domain containing 1 (220945_x_at), score: 0.74 MAP1Amicrotubule-associated protein 1A (203151_at), score: 0.63 ME1malic enzyme 1, NADP(+)-dependent, cytosolic (204059_s_at), score: 0.61 ME3malic enzyme 3, NADP(+)-dependent, mitochondrial (204663_at), score: 0.6 MEFVMediterranean fever (208262_x_at), score: -0.6 MICAL2microtubule associated monoxygenase, calponin and LIM domain containing 2 (212473_s_at), score: 0.63 MLF1myeloid leukemia factor 1 (204784_s_at), score: -0.57 MLLT3myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 (204918_s_at), score: 0.6 MMDmonocyte to macrophage differentiation-associated (203414_at), score: -0.67 MOXD1monooxygenase, DBH-like 1 (209708_at), score: 0.64 MTUS1mitochondrial tumor suppressor 1 (212096_s_at), score: -0.6 MYL9myosin, light chain 9, regulatory (201058_s_at), score: 0.59 MYO1Bmyosin IB (212364_at), score: 0.69 MYO1Emyosin IE (203072_at), score: 0.65 NAV3neuron navigator 3 (204823_at), score: 0.77 NBEAneurobeachin (221207_s_at), score: 0.85 NBPF10neuroblastoma breakpoint family, member 10 (214693_x_at), score: -0.54 NEDD9neural precursor cell expressed, developmentally down-regulated 9 (202149_at), score: 0.6 NESnestin (218678_at), score: 0.81 NOTCH3Notch homolog 3 (Drosophila) (203238_s_at), score: 0.61 NT5E5'-nucleotidase, ecto (CD73) (203939_at), score: 0.61 OPTNoptineurin (202073_at), score: 0.61 OXTRoxytocin receptor (206825_at), score: 0.64 PCGF1polycomb group ring finger 1 (210023_s_at), score: -0.52 PDE8Aphosphodiesterase 8A (212522_at), score: -0.53 PDGFRAplatelet-derived growth factor receptor, alpha polypeptide (203131_at), score: 0.69 PDLIM1PDZ and LIM domain 1 (208690_s_at), score: 0.72 PER2period homolog 2 (Drosophila) (205251_at), score: -0.54 PEX5peroxisomal biogenesis factor 5 (203244_at), score: -0.64 PIP4K2Cphosphatidylinositol-5-phosphate 4-kinase, type II, gamma (218942_at), score: -0.53 PLCB1phospholipase C, beta 1 (phosphoinositide-specific) (213222_at), score: 0.66 PLK2polo-like kinase 2 (Drosophila) (201939_at), score: -0.63 PMS2CLPMS2 C-terminal like pseudogene (221206_at), score: -0.55 PPARGC1Aperoxisome proliferator-activated receptor gamma, coactivator 1 alpha (219195_at), score: -0.68 PRSS2protease, serine, 2 (trypsin 2) (205402_x_at), score: 0.68 PRSS3protease, serine, 3 (213421_x_at), score: 0.66 PTPN13protein tyrosine phosphatase, non-receptor type 13 (APO-1/CD95 (Fas)-associated phosphatase) (204201_s_at), score: 0.62 RGNEFRho-guanine nucleotide exchange factor (219610_at), score: -0.65 RPL5ribosomal protein L5 (213689_x_at), score: 0.63 S100A4S100 calcium binding protein A4 (203186_s_at), score: 0.67 SAP30BPSAP30 binding protein (217965_s_at), score: -0.54 SAPS2SAPS domain family, member 2 (202792_s_at), score: -0.55 SAT1spermidine/spermine N1-acetyltransferase 1 (213988_s_at), score: -0.57 SCD5stearoyl-CoA desaturase 5 (220232_at), score: -0.55 SCN3Asodium channel, voltage-gated, type III, alpha subunit (210432_s_at), score: 0.77 SCN9Asodium channel, voltage-gated, type IX, alpha subunit (206950_at), score: 0.64 SEPP1selenoprotein P, plasma, 1 (201427_s_at), score: -0.57 SH3BGRL3SH3 domain binding glutamic acid-rich protein like 3 (221269_s_at), score: 0.63 SKP1S-phase kinase-associated protein 1 (200719_at), score: -0.56 SLC25A37solute carrier family 25, member 37 (218136_s_at), score: -0.6 SLC39A8solute carrier family 39 (zinc transporter), member 8 (209267_s_at), score: -0.56 SMA5glucuronidase, beta pseudogene (215043_s_at), score: -0.53 SNNstannin (218032_at), score: -0.72 SOX9SRY (sex determining region Y)-box 9 (202935_s_at), score: -0.57 SPATA2spermatogenesis associated 2 (204433_s_at), score: -0.53 SPTLC3serine palmitoyltransferase, long chain base subunit 3 (220456_at), score: -0.53 ST6GAL1ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 (201998_at), score: 0.68 STAT6signal transducer and activator of transcription 6, interleukin-4 induced (201331_s_at), score: 0.73 SYNGR1synaptogyrin 1 (210613_s_at), score: 0.63 SYNMsynemin, intermediate filament protein (212730_at), score: 0.89 SYT11synaptotagmin XI (209198_s_at), score: 0.71 TAGLNtransgelin (205547_s_at), score: 0.61 TBC1D2BTBC1 domain family, member 2B (212796_s_at), score: 0.6 TBC1D8TBC1 domain family, member 8 (with GRAM domain) (204526_s_at), score: -0.58 TBXA2Rthromboxane A2 receptor (336_at), score: 1 TCF7transcription factor 7 (T-cell specific, HMG-box) (205255_x_at), score: -0.53 TEAD3TEA domain family member 3 (209454_s_at), score: 0.72 TFDP2transcription factor Dp-2 (E2F dimerization partner 2) (203588_s_at), score: -0.58 TGFB1I1transforming growth factor beta 1 induced transcript 1 (209651_at), score: 0.68 THSD1thrombospondin, type I, domain containing 1 (219477_s_at), score: -0.68 TIMM22translocase of inner mitochondrial membrane 22 homolog (yeast) (219184_x_at), score: 0.61 TIMM8Atranslocase of inner mitochondrial membrane 8 homolog A (yeast) (210800_at), score: -0.53 TMEM111transmembrane protein 111 (217882_at), score: -0.59 TMEM2transmembrane protein 2 (218113_at), score: 0.62 TMEM209transmembrane protein 209 (222267_at), score: -0.56 TNFSF4tumor necrosis factor (ligand) superfamily, member 4 (207426_s_at), score: 0.73 TNS1tensin 1 (221748_s_at), score: 0.67 TRAF5TNF receptor-associated factor 5 (204352_at), score: 0.67 TRY6trypsinogen C (215395_x_at), score: 0.71 UGDHUDP-glucose dehydrogenase (203343_at), score: 0.69 UGP2UDP-glucose pyrophosphorylase 2 (205480_s_at), score: 0.59 VGLL3vestigial like 3 (Drosophila) (220327_at), score: 0.67 WASLWiskott-Aldrich syndrome-like (205809_s_at), score: -0.53 WWP2WW domain containing E3 ubiquitin protein ligase 2 (204022_at), score: 0.61 ZMIZ1zinc finger, MIZ-type containing 1 (212124_at), score: -0.61 ZNF212zinc finger protein 212 (203985_at), score: -0.52 ZNF273zinc finger protein 273 (215239_x_at), score: -0.57 ZYXzyxin (200808_s_at), score: 0.61
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-4219-raw-cel-1311956275.cel | 8 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956398.cel | 12 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956457.cel | 14 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-3860-raw-cel-1561690231.cel | 4 | 5 | HGPS | hgu133a | HGPS | AG10750 |
| E-GEOD-3860-raw-cel-1561690432.cel | 16 | 5 | HGPS | hgu133a | HGPS | AG10750 |
| E-GEOD-3860-raw-cel-1561690336.cel | 9 | 5 | HGPS | hgu133a | HGPS | AG10750 |
| E-GEOD-4219-raw-cel-1311956358.cel | 10 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956178.cel | 6 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956824.cel | 24 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |