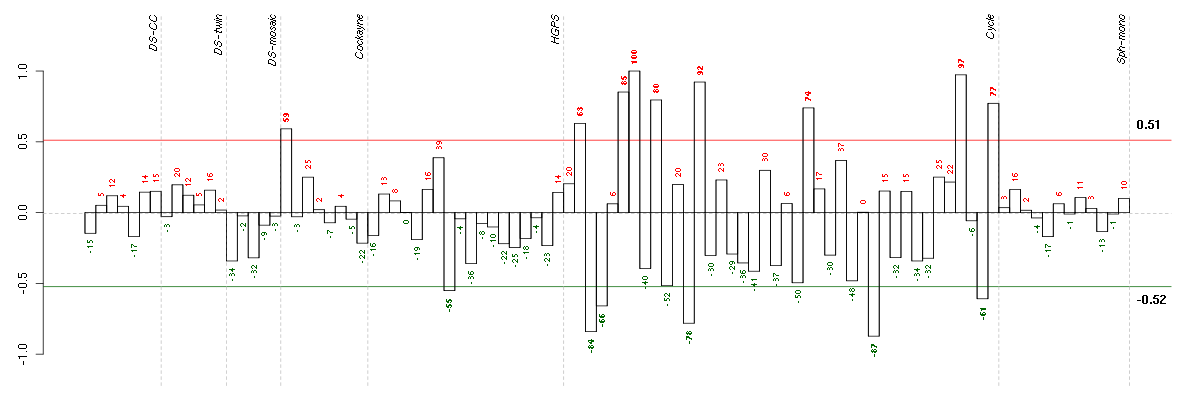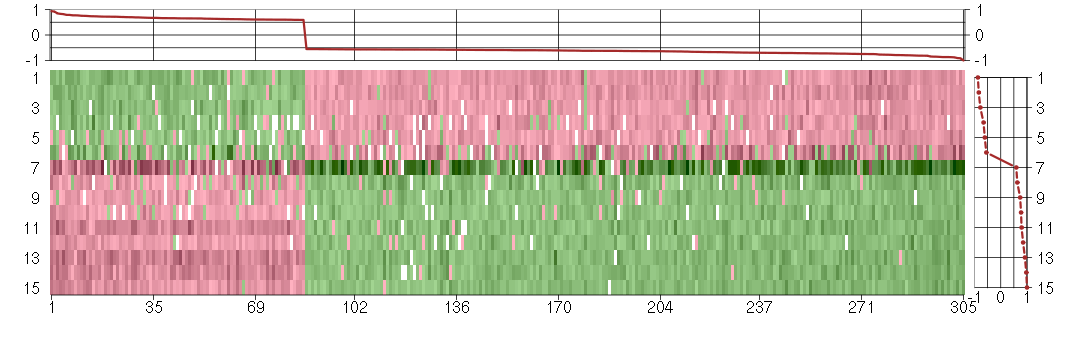

Under-expression is coded with green,
over-expression with red color.

MAPKKK cascade
Cascade of at least three protein kinase activities culminating in the phosphorylation and activation of a MAP kinase. MAPKKK cascades lie downstream of numerous signaling pathways.
activation of MAPK activity
The initiation of the activity of the inactive enzyme MAP kinase by phosphorylation by a MAPKK.
metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
signal transduction
The cascade of processes by which a signal interacts with a receptor, causing a change in the level or activity of a second messenger or other downstream target, and ultimately effecting a change in the functioning of the cell.
protein modification process
The covalent alteration of one or more amino acids occurring in proteins, peptides and nascent polypeptides (co-translational, post-translational modifications). Includes the modification of charged tRNAs that are destined to occur in a protein (pre-translation modification).
protein amino acid phosphorylation
The process of introducing a phosphate group on to a protein.
phosphorus metabolic process
The chemical reactions and pathways involving the nonmetallic element phosphorus or compounds that contain phosphorus, usually in the form of a phosphate group (PO4).
phosphate metabolic process
The chemical reactions and pathways involving the phosphate group, the anion or salt of any phosphoric acid.
response to stress
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a disturbance in organismal or cellular homeostasis, usually, but not necessarily, exogenous (e.g. temperature, humidity, ionizing radiation).
cell communication
Any process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
intracellular signaling cascade
A series of reactions within the cell that occur as a result of a single trigger reaction or compound.
protein kinase cascade
A series of reactions, mediated by protein kinases, which occurs as a result of a single trigger reaction or compound.
JNK cascade
A cascade of protein kinase activities, culminating in the phosphorylation and activation of a member of the JUN kinase subfamily of stress-activated protein kinases, which in turn are a subfamily of mitogen-activated protein (MAP) kinases that is activated primarily by cytokines and exposure to environmental stress.
activation of JUN kinase activity
The initiation of the activity of the inactive enzyme JUN kinase by phosphorylation by a JUN kinase kinase (JNKK).
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
phosphorylation
The process of introducing a phosphate group into a molecule, usually with the formation of a phosphoric ester, a phosphoric anhydride or a phosphoric amide.
protein metabolic process
The chemical reactions and pathways involving a specific protein, rather than of proteins in general. Includes protein modification.
stress-activated protein kinase signaling pathway
A series of molecular signals in which a stress-activated protein kinase (SAPK) cascade relays one or more of the signals.
cellular response to stress
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating the organism is under stress. The stress is usually, but not necessarily, exogenous (e.g. temperature, humidity, ionizing radiation).
positive regulation of kinase activity
Any process that activates or increases the frequency, rate or extent of kinase activity, the catalysis of the transfer of a phosphate group, usually from ATP, to a substrate molecule.
cellular biopolymer metabolic process
The chemical reactions and pathways involving biopolymers, long, repeating chains of monomers found in nature, such as polysaccharides and proteins, as carried out by individual cells.
positive regulation of catalytic activity
Any process that activates or increases the activity of an enzyme.
macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
biopolymer metabolic process
The chemical reactions and pathways involving biopolymers, long, repeating chains of monomers found in nature, such as polysaccharides and proteins.
regulation of MAP kinase activity
Any process that modulates the frequency, rate or extent of MAP kinase activity.
positive regulation of MAP kinase activity
Any process that activates or increases the frequency, rate or extent of MAP kinase activity.
biopolymer modification
The covalent alteration of one or more monomeric units in a polypeptide, polynucleotide, polysaccharide, or other biological polymer, resulting in a change in its properties.
regulation of JUN kinase activity
Any process that modulates the frequency, rate or extent of JUN kinase activity.
positive regulation of JUN kinase activity
Any process that activates or increases the frequency, rate or extent of JUN kinase activity.
regulation of kinase activity
Any process that modulates the frequency, rate or extent of kinase activity, the catalysis of the transfer of a phosphate group, usually from ATP, to a substrate molecule.
post-translational protein modification
The covalent alteration of one or more amino acids occurring in a protein after the protein has been completely translated and released from the ribosome.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
primary metabolic process
The chemical reactions and pathways involving those compounds which are formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
cellular protein metabolic process
The chemical reactions and pathways involving a specific protein, rather than of proteins in general, occurring at the level of an individual cell. Includes protein modification.
regulation of protein kinase activity
Any process that modulates the frequency, rate or extent of protein kinase activity.
positive regulation of protein kinase activity
Any process that activates or increases the frequency, rate or extent of protein kinase activity.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of catalytic activity
Any process that modulates the activity of an enzyme.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
response to stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
regulation of transferase activity
Any process that modulates the frequency, rate or extent of transferase activity, the catalysis of the transfer of a group, e.g. a methyl group, glycosyl group, acyl group, phosphorus-containing, or other groups, from one compound (generally regarded as the donor) to another compound (generally regarded as the acceptor). Transferase is the systematic name for any enzyme of EC class 2.
positive regulation of transferase activity
Any process that activates or increases the frequency, rate or extent of transferase activity, the catalysis of the transfer of a group, e.g. a methyl group, glycosyl group, acyl group, phosphorus-containing, or other groups, from a donor compound to an acceptor.
cellular response to stimulus
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
regulation of molecular function
Any process that modulates the frequency, rate or extent of molecular functions. Molecular functions are elemental biological activities occurring at the molecular level, such as catalysis or binding.
all
This term is the most general term possible
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
cellular response to stimulus
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
signal transduction
The cascade of processes by which a signal interacts with a receptor, causing a change in the level or activity of a second messenger or other downstream target, and ultimately effecting a change in the functioning of the cell.
cellular response to stress
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating the organism is under stress. The stress is usually, but not necessarily, exogenous (e.g. temperature, humidity, ionizing radiation).
protein metabolic process
The chemical reactions and pathways involving a specific protein, rather than of proteins in general. Includes protein modification.
cellular biopolymer metabolic process
The chemical reactions and pathways involving biopolymers, long, repeating chains of monomers found in nature, such as polysaccharides and proteins, as carried out by individual cells.
cellular protein metabolic process
The chemical reactions and pathways involving a specific protein, rather than of proteins in general, occurring at the level of an individual cell. Includes protein modification.
protein modification process
The covalent alteration of one or more amino acids occurring in proteins, peptides and nascent polypeptides (co-translational, post-translational modifications). Includes the modification of charged tRNAs that are destined to occur in a protein (pre-translation modification).
stress-activated protein kinase signaling pathway
A series of molecular signals in which a stress-activated protein kinase (SAPK) cascade relays one or more of the signals.
positive regulation of transferase activity
Any process that activates or increases the frequency, rate or extent of transferase activity, the catalysis of the transfer of a group, e.g. a methyl group, glycosyl group, acyl group, phosphorus-containing, or other groups, from a donor compound to an acceptor.
positive regulation of kinase activity
Any process that activates or increases the frequency, rate or extent of kinase activity, the catalysis of the transfer of a phosphate group, usually from ATP, to a substrate molecule.
protein amino acid phosphorylation
The process of introducing a phosphate group on to a protein.
activation of MAPK activity
The initiation of the activity of the inactive enzyme MAP kinase by phosphorylation by a MAPKK.
JNK cascade
A cascade of protein kinase activities, culminating in the phosphorylation and activation of a member of the JUN kinase subfamily of stress-activated protein kinases, which in turn are a subfamily of mitogen-activated protein (MAP) kinases that is activated primarily by cytokines and exposure to environmental stress.
positive regulation of protein kinase activity
Any process that activates or increases the frequency, rate or extent of protein kinase activity.
activation of JUN kinase activity
The initiation of the activity of the inactive enzyme JUN kinase by phosphorylation by a JUN kinase kinase (JNKK).
positive regulation of MAP kinase activity
Any process that activates or increases the frequency, rate or extent of MAP kinase activity.
activation of MAPK activity
The initiation of the activity of the inactive enzyme MAP kinase by phosphorylation by a MAPKK.
positive regulation of JUN kinase activity
Any process that activates or increases the frequency, rate or extent of JUN kinase activity.
activation of JUN kinase activity
The initiation of the activity of the inactive enzyme JUN kinase by phosphorylation by a JUN kinase kinase (JNKK).


AAASachalasia, adrenocortical insufficiency, alacrimia (Allgrove, triple-A) (218075_at), score: -0.64 ABCD1ATP-binding cassette, sub-family D (ALD), member 1 (205142_x_at), score: -0.77 ABLIM1actin binding LIM protein 1 (200965_s_at), score: -0.57 ACSL4acyl-CoA synthetase long-chain family member 4 (202422_s_at), score: 0.65 ACTN3actinin, alpha 3 (206891_at), score: 0.59 ACTR3BARP3 actin-related protein 3 homolog B (yeast) (218868_at), score: 0.7 ADARadenosine deaminase, RNA-specific (201786_s_at), score: -0.59 ADCK2aarF domain containing kinase 2 (221893_s_at), score: -0.67 ADCY3adenylate cyclase 3 (209321_s_at), score: -0.58 AGPAT11-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) (215535_s_at), score: -0.7 AGRNagrin (212285_s_at), score: -0.56 AHDC1AT hook, DNA binding motif, containing 1 (205002_at), score: -0.56 ALDH1L1aldehyde dehydrogenase 1 family, member L1 (215798_at), score: 0.72 ANXA2P1annexin A2 pseudogene 1 (210876_at), score: 0.61 AP2A2adaptor-related protein complex 2, alpha 2 subunit (212159_x_at), score: -0.58 APPamyloid beta (A4) precursor protein (200602_at), score: 0.63 ARFGAP3ADP-ribosylation factor GTPase activating protein 3 (202211_at), score: 0.61 ARSAarylsulfatase A (204443_at), score: -0.68 ATF5activating transcription factor 5 (204999_s_at), score: -0.62 ATN1atrophin 1 (40489_at), score: -0.77 ATP2C1ATPase, Ca++ transporting, type 2C, member 1 (211137_s_at), score: 0.73 B3GAT3beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) (203452_at), score: -0.72 BAHCC1BAH domain and coiled-coil containing 1 (219218_at), score: -0.58 BAP1BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) (201419_at), score: -0.72 BAT2HLA-B associated transcript 2 (212081_x_at), score: -0.81 BAT2LHLA-B associated transcript 2-like (212068_s_at), score: -0.71 BIN1bridging integrator 1 (210201_x_at), score: -0.62 BRD4bromodomain containing 4 (202102_s_at), score: -0.6 BTBD2BTB (POZ) domain containing 2 (207722_s_at), score: -0.8 BTG2BTG family, member 2 (201236_s_at), score: -0.56 C10orf97chromosome 10 open reading frame 97 (218297_at), score: 0.63 C15orf33chromosome 15 open reading frame 33 (216411_s_at), score: 0.65 C16orf45chromosome 16 open reading frame 45 (212736_at), score: -0.63 C16orf57chromosome 16 open reading frame 57 (218060_s_at), score: -0.62 C20orf20chromosome 20 open reading frame 20 (218586_at), score: -0.63 C4orf43chromosome 4 open reading frame 43 (218513_at), score: 0.72 C5orf44chromosome 5 open reading frame 44 (218674_at), score: 0.67 CABIN1calcineurin binding protein 1 (37652_at), score: -0.67 CALB2calbindin 2 (205428_s_at), score: 0.83 CALCOCO1calcium binding and coiled-coil domain 1 (209002_s_at), score: -0.64 CAPN5calpain 5 (205166_at), score: -0.6 CDC25Bcell division cycle 25 homolog B (S. pombe) (201853_s_at), score: -0.62 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (217849_s_at), score: -0.87 CDC42EP1CDC42 effector protein (Rho GTPase binding) 1 (204693_at), score: -0.68 CDC42EP4CDC42 effector protein (Rho GTPase binding) 4 (218062_x_at), score: -0.75 CDH6cadherin 6, type 2, K-cadherin (fetal kidney) (205532_s_at), score: 0.6 CHMP1Achromatin modifying protein 1A (201933_at), score: -0.56 CICcapicua homolog (Drosophila) (212784_at), score: -0.87 CITcitron (rho-interacting, serine/threonine kinase 21) (212801_at), score: -0.6 CIZ1CDKN1A interacting zinc finger protein 1 (205516_x_at), score: -0.81 CLIP2CAP-GLY domain containing linker protein 2 (211031_s_at), score: -0.61 CNIH3cornichon homolog 3 (Drosophila) (214841_at), score: -0.67 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: -0.79 COL18A1collagen, type XVIII, alpha 1 (209081_s_at), score: -0.59 CORINcorin, serine peptidase (220356_at), score: 0.62 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: -0.87 CScitrate synthase (208660_at), score: -0.73 CUL7cullin 7 (36084_at), score: -0.56 DAXXdeath-domain associated protein (201763_s_at), score: -0.59 DENND2ADENN/MADD domain containing 2A (53991_at), score: -0.57 DENND3DENN/MADD domain containing 3 (212974_at), score: -0.6 DENND5BDENN/MADD domain containing 5B (215058_at), score: 0.62 DERL2Der1-like domain family, member 2 (218333_at), score: 0.59 DKFZP564O0823DKFZP564O0823 protein (204687_at), score: 0.6 DMBT1deleted in malignant brain tumors 1 (208250_s_at), score: 0.62 DMPKdystrophia myotonica-protein kinase (37996_s_at), score: -0.72 DMWDdystrophia myotonica, WD repeat containing (33768_at), score: -0.69 DNAJB9DnaJ (Hsp40) homolog, subfamily B, member 9 (202843_at), score: 0.69 DNAJC6DnaJ (Hsp40) homolog, subfamily C, member 6 (204720_s_at), score: 0.64 DNM2dynamin 2 (202253_s_at), score: -0.64 DOK4docking protein 4 (209691_s_at), score: -0.57 DOPEY1dopey family member 1 (40612_at), score: 0.64 ECM1extracellular matrix protein 1 (209365_s_at), score: -0.56 EHBP1L1EH domain binding protein 1-like 1 (221755_at), score: -0.59 EHMT2euchromatic histone-lysine N-methyltransferase 2 (202326_at), score: -0.56 ENTPD1ectonucleoside triphosphate diphosphohydrolase 1 (209473_at), score: 0.65 EPHA4EPH receptor A4 (206114_at), score: 0.75 ERBB2IPerbb2 interacting protein (217941_s_at), score: 0.71 EVLEnah/Vasp-like (217838_s_at), score: -0.58 FAM18Bfamily with sequence similarity 18, member B (218446_s_at), score: 0.72 FAM62Afamily with sequence similarity 62 (C2 domain containing), member A (208858_s_at), score: -0.59 FAT4FAT tumor suppressor homolog 4 (Drosophila) (219427_at), score: 0.66 FBXL11F-box and leucine-rich repeat protein 11 (208989_s_at), score: -0.65 FBXL14F-box and leucine-rich repeat protein 14 (213145_at), score: -0.81 FBXO17F-box protein 17 (220233_at), score: -0.68 FCER1GFc fragment of IgE, high affinity I, receptor for; gamma polypeptide (204232_at), score: 0.64 FCF1FCF1 small subunit (SSU) processome component homolog (S. cerevisiae) (219927_at), score: 0.68 FHOD1formin homology 2 domain containing 1 (218530_at), score: -0.62 FKBP8FK506 binding protein 8, 38kDa (208255_s_at), score: -0.56 FKRPfukutin related protein (219853_at), score: -0.57 FLJ12529pre-mRNA cleavage factor I, 59 kDa subunit (217866_at), score: -0.72 FOSL1FOS-like antigen 1 (204420_at), score: -0.58 FOXC2forkhead box C2 (MFH-1, mesenchyme forkhead 1) (214520_at), score: -0.67 FOXF1forkhead box F1 (205935_at), score: -0.61 FOXK2forkhead box K2 (203064_s_at), score: -0.91 GABARAPL3GABA(A) receptors associated protein like 3 (pseudogene) (211458_s_at), score: 0.66 GK3Pglycerol kinase 3 pseudogene (215966_x_at), score: 0.65 GMCL1germ cell-less homolog 1 (Drosophila) (218458_at), score: 0.6 GMPRguanosine monophosphate reductase (204187_at), score: -0.71 GOLT1Bgolgi transport 1 homolog B (S. cerevisiae) (218193_s_at), score: 0.61 GRINAglutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) (212090_at), score: -0.62 H1FXH1 histone family, member X (204805_s_at), score: -0.73 HAB1B1 for mucin (215778_x_at), score: 0.7 HBS1LHBS1-like (S. cerevisiae) (209314_s_at), score: 0.59 HECTD3HECT domain containing 3 (218632_at), score: -0.57 HLA-DQB1major histocompatibility complex, class II, DQ beta 1 (211654_x_at), score: 0.6 HMGA1high mobility group AT-hook 1 (210457_x_at), score: -0.78 HNRNPH2heterogeneous nuclear ribonucleoprotein H2 (H') (201132_at), score: 0.62 HNRNPUL1heterogeneous nuclear ribonucleoprotein U-like 1 (209675_s_at), score: -0.73 HSD17B6hydroxysteroid (17-beta) dehydrogenase 6 homolog (mouse) (37512_at), score: 0.79 HSF1heat shock transcription factor 1 (202344_at), score: -0.64 HSPA12Aheat shock 70kDa protein 12A (214434_at), score: -0.71 IDUAiduronidase, alpha-L- (205059_s_at), score: -0.79 IL9Rinterleukin 9 receptor (217212_s_at), score: 0.71 INTS1integrator complex subunit 1 (212212_s_at), score: -0.63 ITGA1integrin, alpha 1 (214660_at), score: 0.66 JUNDjun D proto-oncogene (203751_x_at), score: -0.72 JUPjunction plakoglobin (201015_s_at), score: -0.84 KAL1Kallmann syndrome 1 sequence (205206_at), score: 0.85 KCNJ8potassium inwardly-rectifying channel, subfamily J, member 8 (205303_at), score: 0.6 KCTD13potassium channel tetramerisation domain containing 13 (45653_at), score: -0.56 KCTD2potassium channel tetramerisation domain containing 2 (212564_at), score: -0.6 KIAA1305KIAA1305 (220911_s_at), score: -0.71 KLHL23kelch-like 23 (Drosophila) (213610_s_at), score: 0.74 KPNA6karyopherin alpha 6 (importin alpha 7) (212101_at), score: -0.6 LEPREL2leprecan-like 2 (204854_at), score: -0.7 LLGL2lethal giant larvae homolog 2 (Drosophila) (203713_s_at), score: 0.61 LMF2lipase maturation factor 2 (212682_s_at), score: -0.64 LOC100132540similar to LOC339047 protein (214870_x_at), score: -0.58 LOC339047hypothetical protein LOC339047 (221501_x_at), score: -0.68 LOC391132similar to hCG2041276 (216177_at), score: 0.65 LOC90379hypothetical protein BC002926 (221849_s_at), score: -0.59 LPGAT1lysophosphatidylglycerol acyltransferase 1 (202651_at), score: 0.66 LRDDleucine-rich repeats and death domain containing (221640_s_at), score: -0.74 LRP2low density lipoprotein-related protein 2 (205710_at), score: -0.59 LRP8low density lipoprotein receptor-related protein 8, apolipoprotein e receptor (205282_at), score: 0.65 LRRN3leucine rich repeat neuronal 3 (209841_s_at), score: 0.62 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: -0.91 MAP2K3mitogen-activated protein kinase kinase 3 (207667_s_at), score: -0.56 MAP3K11mitogen-activated protein kinase kinase kinase 11 (203652_at), score: -0.6 MAP3K6mitogen-activated protein kinase kinase kinase 6 (219278_at), score: -0.8 MAP4microtubule-associated protein 4 (200836_s_at), score: -0.61 MAP7D1MAP7 domain containing 1 (217943_s_at), score: -0.69 MAST2microtubule associated serine/threonine kinase 2 (211593_s_at), score: -0.72 MAST4microtubule associated serine/threonine kinase family member 4 (40016_g_at), score: -0.61 MED15mediator complex subunit 15 (222175_s_at), score: -0.59 MFAP3Lmicrofibrillar-associated protein 3-like (205442_at), score: 0.75 MGAT1mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase (201126_s_at), score: -0.61 MINK1misshapen-like kinase 1 (zebrafish) (214246_x_at), score: -0.63 MLL4myeloid/lymphoid or mixed-lineage leukemia 4 (203419_at), score: -0.63 MMP10matrix metallopeptidase 10 (stromelysin 2) (205680_at), score: 0.65 MPP1membrane protein, palmitoylated 1, 55kDa (202974_at), score: -0.57 MUC1mucin 1, cell surface associated (207847_s_at), score: -0.6 MUL1mitochondrial E3 ubiquitin ligase 1 (218246_at), score: -0.7 MYL4myosin, light chain 4, alkali; atrial, embryonic (210088_x_at), score: 0.68 MYO1Bmyosin IB (212364_at), score: 0.63 NARG1LNMDA receptor regulated 1-like (219378_at), score: 0.65 NBL1neuroblastoma, suppression of tumorigenicity 1 (37005_at), score: -0.62 NCDNneurochondrin (209556_at), score: -0.58 NCK2NCK adaptor protein 2 (203315_at), score: -0.58 NCKAP1NCK-associated protein 1 (207738_s_at), score: 0.67 NCOR2nuclear receptor co-repressor 2 (207760_s_at), score: -0.57 NDPNorrie disease (pseudoglioma) (206022_at), score: -0.77 NEFLneurofilament, light polypeptide (221805_at), score: 0.62 NEFMneurofilament, medium polypeptide (205113_at), score: 0.94 NF1neurofibromin 1 (211094_s_at), score: -0.62 NFATC4nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 (205897_at), score: -0.63 NFKBIAnuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha (201502_s_at), score: -0.66 NINJ1ninjurin 1 (203045_at), score: -0.6 NPDC1neural proliferation, differentiation and control, 1 (218086_at), score: -0.57 NPIPnuclear pore complex interacting protein (204538_x_at), score: -0.74 OAS32'-5'-oligoadenylate synthetase 3, 100kDa (218400_at), score: -0.6 OBFC2Boligonucleotide/oligosaccharide-binding fold containing 2B (218903_s_at), score: -0.57 OGDHoxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) (201282_at), score: -0.57 OLFML2Bolfactomedin-like 2B (213125_at), score: -0.75 OLR1oxidized low density lipoprotein (lectin-like) receptor 1 (210004_at), score: 0.63 OPCMLopioid binding protein/cell adhesion molecule-like (214111_at), score: 0.6 OSTM1osteopetrosis associated transmembrane protein 1 (218196_at), score: 0.7 PAK1p21 protein (Cdc42/Rac)-activated kinase 1 (209615_s_at), score: -0.66 PARNpoly(A)-specific ribonuclease (deadenylation nuclease) (203905_at), score: -0.57 PARVBparvin, beta (37965_at), score: -0.79 PCDH9protocadherin 9 (219737_s_at), score: 0.82 PCDHG@protocadherin gamma cluster (215836_s_at), score: -0.68 PCDHGA1protocadherin gamma subfamily A, 1 (209079_x_at), score: -0.73 PCDHGA11protocadherin gamma subfamily A, 11 (211876_x_at), score: -0.56 PCNXpecanex homolog (Drosophila) (213159_at), score: -0.69 PCYT2phosphate cytidylyltransferase 2, ethanolamine (209577_at), score: -0.61 PDE5Aphosphodiesterase 5A, cGMP-specific (206757_at), score: -0.62 PDE7Bphosphodiesterase 7B (220343_at), score: 0.67 PEX6peroxisomal biogenesis factor 6 (204545_at), score: -0.67 PEX7peroxisomal biogenesis factor 7 (205420_at), score: 0.61 PHF7PHD finger protein 7 (215622_x_at), score: 0.73 PIGOphosphatidylinositol glycan anchor biosynthesis, class O (209998_at), score: -0.56 PIP5K1Cphosphatidylinositol-4-phosphate 5-kinase, type I, gamma (212518_at), score: -0.88 PKN1protein kinase N1 (202161_at), score: -0.75 PLAUplasminogen activator, urokinase (211668_s_at), score: -0.69 PLK1polo-like kinase 1 (Drosophila) (202240_at), score: -0.57 PNMAL1PNMA-like 1 (218824_at), score: -0.57 PNPLA2patatin-like phospholipase domain containing 2 (212705_x_at), score: -0.57 PNPLA6patatin-like phospholipase domain containing 6 (203718_at), score: -0.64 POLR2Apolymerase (RNA) II (DNA directed) polypeptide A, 220kDa (202725_at), score: -0.77 POLSpolymerase (DNA directed) sigma (202466_at), score: -0.62 POM121POM121 membrane glycoprotein (rat) (212178_s_at), score: -0.7 POM121CPOM121 membrane glycoprotein C (213360_s_at), score: -0.55 POMZP3POM (POM121 homolog, rat) and ZP3 fusion (204148_s_at), score: -0.74 PRKCDprotein kinase C, delta (202545_at), score: -0.71 PRMT3protein arginine methyltransferase 3 (213320_at), score: 0.72 PTDSS1phosphatidylserine synthase 1 (201433_s_at), score: -0.55 PTOV1prostate tumor overexpressed 1 (212032_s_at), score: -0.55 PTPN9protein tyrosine phosphatase, non-receptor type 9 (202958_at), score: -0.6 PTPROprotein tyrosine phosphatase, receptor type, O (211600_at), score: -0.7 PXNpaxillin (211823_s_at), score: -0.71 RAB1ARAB1A, member RAS oncogene family (213440_at), score: 0.71 RAB22ARAB22A, member RAS oncogene family (218360_at), score: -0.59 RAD54L2RAD54-like 2 (S. cerevisiae) (213205_s_at), score: -0.65 RANBP3RAN binding protein 3 (210120_s_at), score: -0.58 RBM15BRNA binding motif protein 15B (202689_at), score: -0.71 REREarginine-glutamic acid dipeptide (RE) repeats (200940_s_at), score: -0.85 RGS2regulator of G-protein signaling 2, 24kDa (202388_at), score: -0.55 RNF11ring finger protein 11 (208924_at), score: 0.69 RNF123ring finger protein 123 (221063_x_at), score: -0.57 RNF19Bring finger protein 19B (213038_at), score: -0.62 RNF220ring finger protein 220 (219988_s_at), score: -0.74 RNF24ring finger protein 24 (204669_s_at), score: -0.57 RNF40ring finger protein 40 (206845_s_at), score: -0.57 RP5-1000E10.4suppressor of IKK epsilon (221705_s_at), score: 0.68 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: 0.92 RPRD2regulation of nuclear pre-mRNA domain containing 2 (212553_at), score: -0.67 RPS17P5ribosomal protein S17 pseudogene 5 (216348_at), score: 0.79 RPS25ribosomal protein S25 (200091_s_at), score: 0.61 RPS6KA4ribosomal protein S6 kinase, 90kDa, polypeptide 4 (204632_at), score: -0.6 SAP30LSAP30-like (219129_s_at), score: -0.58 SAV1salvador homolog 1 (Drosophila) (218276_s_at), score: -0.65 SBF1SET binding factor 1 (39835_at), score: -0.56 SBNO2strawberry notch homolog 2 (Drosophila) (204166_at), score: -0.73 SCAMP4secretory carrier membrane protein 4 (213244_at), score: -1 SDAD1SDA1 domain containing 1 (218607_s_at), score: -0.63 SDC3syndecan 3 (202898_at), score: -0.57 SEMA3Fsema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F (35666_at), score: 0.63 SERPINB9serpin peptidase inhibitor, clade B (ovalbumin), member 9 (209723_at), score: -0.57 SF1splicing factor 1 (208313_s_at), score: -0.86 SF3A2splicing factor 3a, subunit 2, 66kDa (37462_i_at), score: -0.63 SH3GLB2SH3-domain GRB2-like endophilin B2 (218813_s_at), score: -0.63 SHBSrc homology 2 domain containing adaptor protein B (204657_s_at), score: -0.62 SIGIRRsingle immunoglobulin and toll-interleukin 1 receptor (TIR) domain (52940_at), score: -0.66 SIGLEC6sialic acid binding Ig-like lectin 6 (210796_x_at), score: 0.61 SIN3BSIN3 homolog B, transcription regulator (yeast) (209352_s_at), score: -0.55 SIPA1signal-induced proliferation-associated 1 (204164_at), score: -0.57 SIRT6sirtuin (silent mating type information regulation 2 homolog) 6 (S. cerevisiae) (219613_s_at), score: -0.62 SLC25A13solute carrier family 25, member 13 (citrin) (203775_at), score: 0.6 SLC2A4RGSLC2A4 regulator (218494_s_at), score: -0.55 SLC33A1solute carrier family 33 (acetyl-CoA transporter), member 1 (203165_s_at), score: 0.6 SLC43A3solute carrier family 43, member 3 (213113_s_at), score: -0.56 SLC4A2solute carrier family 4, anion exchanger, member 2 (erythrocyte membrane protein band 3-like 1) (202111_at), score: -0.57 SLC4A7solute carrier family 4, sodium bicarbonate cotransporter, member 7 (209884_s_at), score: 0.66 SLC5A5solute carrier family 5 (sodium iodide symporter), member 5 (211123_at), score: 0.67 SMAD3SMAD family member 3 (218284_at), score: -0.64 SMARCD1SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 (209518_at), score: -0.6 SMG6Smg-6 homolog, nonsense mediated mRNA decay factor (C. elegans) (214940_s_at), score: -0.75 SMG7Smg-7 homolog, nonsense mediated mRNA decay factor (C. elegans) (201794_s_at), score: -0.7 SMN1survival of motor neuron 1, telomeric (203852_s_at), score: 0.59 SMYD5SMYD family member 5 (209516_at), score: -0.58 SNAP25synaptosomal-associated protein, 25kDa (202508_s_at), score: -0.62 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: -0.79 SORBS3sorbin and SH3 domain containing 3 (209253_at), score: -0.6 SPG7spastic paraplegia 7 (pure and complicated autosomal recessive) (202104_s_at), score: -0.66 SSBP3single stranded DNA binding protein 3 (217991_x_at), score: -0.65 ST5suppression of tumorigenicity 5 (202440_s_at), score: -0.57 STIM1stromal interaction molecule 1 (202764_at), score: -0.63 STRN4striatin, calmodulin binding protein 4 (217903_at), score: -0.67 SULT1B1sulfotransferase family, cytosolic, 1B, member 1 (207601_at), score: 0.77 SYNE1spectrin repeat containing, nuclear envelope 1 (209447_at), score: 0.75 TAF13TAF13 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 18kDa (205966_at), score: 0.72 TAPBPTAP binding protein (tapasin) (208829_at), score: -0.81 TENC1tensin like C1 domain containing phosphatase (tensin 2) (212494_at), score: -0.63 TERF2telomeric repeat binding factor 2 (203611_at), score: -0.69 TIAF1TGFB1-induced anti-apoptotic factor 1 (202039_at), score: -0.71 TICAM2toll-like receptor adaptor molecule 2 (214658_at), score: 0.77 TLE3transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) (206472_s_at), score: -0.69 TLE4transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) (204872_at), score: 0.73 TP53tumor protein p53 (201746_at), score: -0.72 TRIM21tripartite motif-containing 21 (204804_at), score: -0.7 TSC2tuberous sclerosis 2 (215735_s_at), score: -0.7 TSKUtsukushin (218245_at), score: -0.8 TSSC4tumor suppressing subtransferable candidate 4 (218612_s_at), score: -0.56 TXLNAtaxilin alpha (212300_at), score: -0.71 UBE2Zubiquitin-conjugating enzyme E2Z (217750_s_at), score: -0.59 UBTD1ubiquitin domain containing 1 (219172_at), score: -0.75 ULK1unc-51-like kinase 1 (C. elegans) (209333_at), score: -0.59 VAT1vesicle amine transport protein 1 homolog (T. californica) (208626_s_at), score: -0.57 VEGFBvascular endothelial growth factor B (203683_s_at), score: -0.87 VPS13Cvacuolar protein sorting 13 homolog C (S. cerevisiae) (218396_at), score: 0.6 VPS37Bvacuolar protein sorting 37 homolog B (S. cerevisiae) (221704_s_at), score: -0.81 WBP2WW domain binding protein 2 (209117_at), score: -0.58 WDR42AWD repeat domain 42A (202249_s_at), score: -0.74 WDR6WD repeat domain 6 (217734_s_at), score: -0.85 XAB2XPA binding protein 2 (218110_at), score: -0.72 YES1v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 (202932_at), score: -0.67 YTHDC2YTH domain containing 2 (213077_at), score: 0.65 ZC3H3zinc finger CCCH-type containing 3 (213445_at), score: -0.6 ZDHHC7zinc finger, DHHC-type containing 7 (218606_at), score: -0.6 ZFHX3zinc finger homeobox 3 (208033_s_at), score: -0.57 ZFP36L1zinc finger protein 36, C3H type-like 1 (211965_at), score: -0.63 ZNF580zinc finger protein 580 (220748_s_at), score: -0.59
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486411.cel | 39 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690304.cel | 8 | 5 | HGPS | hgu133a | none | GMO8398C |
| E-GEOD-3407-raw-cel-1437949557.cel | 1 | 4 | Cockayne | hgu133a | CS | eGFP |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |