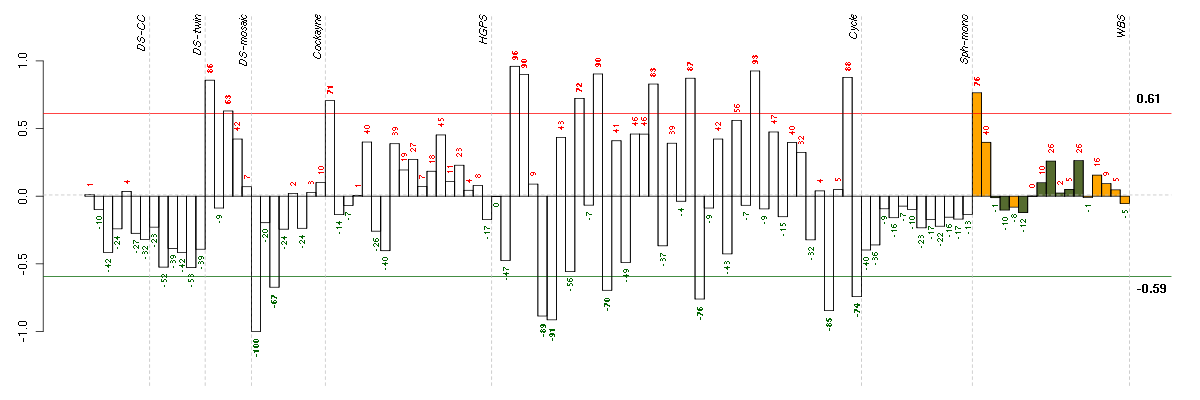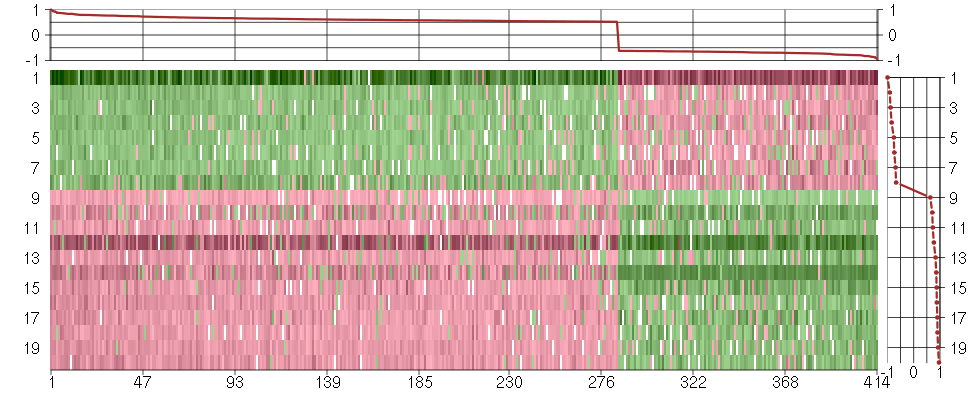

Under-expression is coded with green,
over-expression with red color.

biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of multicellular organismal process
Any process that modulates the frequency, rate or extent of a multicellular organismal process, the processes pertinent to the function of a multicellular organism above the cellular level; includes the integrated processes of tissues and organs.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
all
This term is the most general term possible
regulation of multicellular organismal process
Any process that modulates the frequency, rate or extent of a multicellular organismal process, the processes pertinent to the function of a multicellular organism above the cellular level; includes the integrated processes of tissues and organs.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.


AAASachalasia, adrenocortical insufficiency, alacrimia (Allgrove, triple-A) (218075_at), score: 0.64 ABCD1ATP-binding cassette, sub-family D (ALD), member 1 (205142_x_at), score: 0.74 ACADSBacyl-Coenzyme A dehydrogenase, short/branched chain (205355_at), score: -0.73 ACN9ACN9 homolog (S. cerevisiae) (218981_at), score: -0.64 ADAMTS12ADAM metallopeptidase with thrombospondin type 1 motif, 12 (221421_s_at), score: -0.69 AESamino-terminal enhancer of split (217729_s_at), score: 0.67 AGPAT11-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) (215535_s_at), score: 0.72 AGRNagrin (217419_x_at), score: 0.64 AKAP2A kinase (PRKA) anchor protein 2 (202759_s_at), score: 0.61 AKIRIN1akirin 1 (217893_s_at), score: 0.53 ALDH1L1aldehyde dehydrogenase 1 family, member L1 (215798_at), score: -0.88 ALDH3A1aldehyde dehydrogenase 3 family, memberA1 (205623_at), score: -0.63 ANKFY1ankyrin repeat and FYVE domain containing 1 (219868_s_at), score: 0.64 ANXA2P1annexin A2 pseudogene 1 (210876_at), score: -0.63 AP2A2adaptor-related protein complex 2, alpha 2 subunit (212159_x_at), score: 0.63 APBB1amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) (202652_at), score: 0.64 ARHGDIARho GDP dissociation inhibitor (GDI) alpha (213606_s_at), score: 0.62 ARID1AAT rich interactive domain 1A (SWI-like) (210649_s_at), score: 0.61 ARSAarylsulfatase A (204443_at), score: 0.87 ATF5activating transcription factor 5 (204999_s_at), score: 0.6 ATG9AATG9 autophagy related 9 homolog A (S. cerevisiae) (202492_at), score: 0.58 ATN1atrophin 1 (40489_at), score: 0.61 ATP13A2ATPase type 13A2 (218608_at), score: 0.55 ATP6AP1ATPase, H+ transporting, lysosomal accessory protein 1 (207809_s_at), score: 0.56 B3GAT3beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) (203452_at), score: 0.58 BAIAP3BAI1-associated protein 3 (216356_x_at), score: -0.63 BAP1BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) (201419_at), score: 0.69 BAT2HLA-B associated transcript 2 (212081_x_at), score: 0.84 BAT2LHLA-B associated transcript 2-like (212068_s_at), score: 0.63 BCL2L1BCL2-like 1 (215037_s_at), score: 0.66 BDH13-hydroxybutyrate dehydrogenase, type 1 (211715_s_at), score: -0.71 BRD4bromodomain containing 4 (202102_s_at), score: 0.59 BTBD2BTB (POZ) domain containing 2 (207722_s_at), score: 0.71 C15orf33chromosome 15 open reading frame 33 (216411_s_at), score: -0.72 C16orf45chromosome 16 open reading frame 45 (212736_at), score: 0.56 C19orf22chromosome 19 open reading frame 22 (221764_at), score: 0.67 C1orf109chromosome 1 open reading frame 109 (218712_at), score: -0.64 C1orf69chromosome 1 open reading frame 69 (215490_at), score: -0.68 C20orf149chromosome 20 open reading frame 149 (218010_x_at), score: 0.66 C22orf30chromosome 22 open reading frame 30 (216555_at), score: -0.65 C2orf72chromosome 2 open reading frame 72 (213143_at), score: -0.65 C4orf43chromosome 4 open reading frame 43 (218513_at), score: -0.78 C5orf44chromosome 5 open reading frame 44 (218674_at), score: -0.65 C8orf60chromosome 8 open reading frame 60 (220712_at), score: -0.65 CABIN1calcineurin binding protein 1 (37652_at), score: 0.7 CALB1calbindin 1, 28kDa (205626_s_at), score: -0.62 CALCOCO1calcium binding and coiled-coil domain 1 (209002_s_at), score: 0.68 CAPN1calpain 1, (mu/I) large subunit (200752_s_at), score: 0.62 CCDC41coiled-coil domain containing 41 (219644_at), score: -0.79 CCDC88Acoiled-coil domain containing 88A (219387_at), score: -0.63 CD53CD53 molecule (203416_at), score: -0.63 CDAcytidine deaminase (205627_at), score: 0.56 CDADC1cytidine and dCMP deaminase domain containing 1 (221015_s_at), score: -0.71 CDC23cell division cycle 23 homolog (S. cerevisiae) (202892_at), score: -0.63 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (217849_s_at), score: 0.83 CDC42EP1CDC42 effector protein (Rho GTPase binding) 1 (204693_at), score: 0.76 CENPBcentromere protein B, 80kDa (212437_at), score: 0.59 CHMP1Achromatin modifying protein 1A (201933_at), score: 0.54 CHMP5chromatin modifying protein 5 (218085_at), score: -0.69 CHPFchondroitin polymerizing factor (202175_at), score: 0.63 CHST3carbohydrate (chondroitin 6) sulfotransferase 3 (209834_at), score: 0.56 CICcapicua homolog (Drosophila) (212784_at), score: 0.73 CIZ1CDKN1A interacting zinc finger protein 1 (205516_x_at), score: 0.68 CLDN1claudin 1 (218182_s_at), score: 0.52 CLIP2CAP-GLY domain containing linker protein 2 (211031_s_at), score: 0.57 CLIP3CAP-GLY domain containing linker protein 3 (212358_at), score: 0.6 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: 0.7 CORO1Bcoronin, actin binding protein, 1B (64486_at), score: 0.61 CORO2Bcoronin, actin binding protein, 2B (209789_at), score: 0.52 CPA2carboxypeptidase A2 (pancreatic) (206212_at), score: -0.66 CRCPCGRP receptor component (203898_at), score: -0.69 CRIP2cysteine-rich protein 2 (208978_at), score: 0.6 CST3cystatin C (201360_at), score: 0.54 CTSEcathepsin E (205927_s_at), score: -0.64 CUL7cullin 7 (36084_at), score: 0.63 DBN1drebrin 1 (217025_s_at), score: 0.52 DEAF1deformed epidermal autoregulatory factor 1 (Drosophila) (209407_s_at), score: 0.64 DGCR2DiGeorge syndrome critical region gene 2 (214198_s_at), score: -0.77 DKFZP564O0823DKFZP564O0823 protein (204687_at), score: -0.67 DMPKdystrophia myotonica-protein kinase (37996_s_at), score: 0.6 DNAJC4DnaJ (Hsp40) homolog, subfamily C, member 4 (206782_s_at), score: 0.59 DNM2dynamin 2 (202253_s_at), score: 0.79 DOK4docking protein 4 (209691_s_at), score: 0.64 DOPEY1dopey family member 1 (40612_at), score: -0.83 DRG2developmentally regulated GTP binding protein 2 (203267_s_at), score: 0.74 DSPPdentin sialophosphoprotein (221681_s_at), score: -0.67 DUSP3dual specificity phosphatase 3 (201537_s_at), score: 0.58 DUSP5dual specificity phosphatase 5 (209457_at), score: 0.53 ECE1endothelin converting enzyme 1 (201750_s_at), score: 0.63 ECM1extracellular matrix protein 1 (209365_s_at), score: 0.6 EFHD2EF-hand domain family, member D2 (217992_s_at), score: 0.53 EHBP1L1EH domain binding protein 1-like 1 (221755_at), score: 0.64 EHD2EH-domain containing 2 (221870_at), score: 0.55 ELK1ELK1, member of ETS oncogene family (210376_x_at), score: 0.54 ENGendoglin (201809_s_at), score: 0.53 ENGASEendo-beta-N-acetylglucosaminidase (220349_s_at), score: -0.67 ENOX2ecto-NOX disulfide-thiol exchanger 2 (32042_at), score: -0.64 ENTPD1ectonucleoside triphosphate diphosphohydrolase 1 (209473_at), score: -0.64 EPHA4EPH receptor A4 (206114_at), score: -0.66 EPM2Aepilepsy, progressive myoclonus type 2A, Lafora disease (laforin) (205231_s_at), score: -0.67 EPN2epsin 2 (203463_s_at), score: 0.56 ERBB2v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian) (216836_s_at), score: 0.54 ERBB2IPerbb2 interacting protein (217941_s_at), score: -0.65 EVLEnah/Vasp-like (217838_s_at), score: 0.58 F11coagulation factor XI (206610_s_at), score: -0.66 F8A1coagulation factor VIII-associated (intronic transcript) 1 (203274_at), score: 0.6 FAM176Bfamily with sequence similarity 176, member B (220134_x_at), score: 0.56 FAM62Afamily with sequence similarity 62 (C2 domain containing), member A (208858_s_at), score: 0.53 FAM65Afamily with sequence similarity 65, member A (45749_at), score: 0.65 FBXL18F-box and leucine-rich repeat protein 18 (215068_s_at), score: 0.57 FBXO11F-box protein 11 (203255_at), score: -0.66 FBXO17F-box protein 17 (220233_at), score: 0.63 FBXO2F-box protein 2 (219305_x_at), score: -0.78 FCER1GFc fragment of IgE, high affinity I, receptor for; gamma polypeptide (204232_at), score: -0.79 FCF1FCF1 small subunit (SSU) processome component homolog (S. cerevisiae) (219927_at), score: -0.69 FETUBfetuin B (214417_s_at), score: -0.77 FGF12fibroblast growth factor 12 (207501_s_at), score: -0.71 FGL1fibrinogen-like 1 (205305_at), score: -0.68 FHOD1formin homology 2 domain containing 1 (218530_at), score: 0.73 FKBP8FK506 binding protein 8, 38kDa (208255_s_at), score: 0.78 FKSG2apoptosis inhibitor (208588_at), score: -0.71 FLJ20674hypothetical protein FLJ20674 (220137_at), score: -0.69 FLNBfilamin B, beta (actin binding protein 278) (208613_s_at), score: 0.6 FOXC2forkhead box C2 (MFH-1, mesenchyme forkhead 1) (214520_at), score: 0.76 FOXK2forkhead box K2 (203064_s_at), score: 0.7 FOXO3forkhead box O3 (204132_s_at), score: 0.61 FURINfurin (paired basic amino acid cleaving enzyme) (201945_at), score: 0.65 FUT2fucosyltransferase 2 (secretor status included) (210608_s_at), score: -0.72 GBF1golgi-specific brefeldin A resistant guanine nucleotide exchange factor 1 (201439_at), score: 0.54 GLI2GLI family zinc finger 2 (207034_s_at), score: 0.54 GLUD2glutamate dehydrogenase 2 (215794_x_at), score: 0.55 GNAI2guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2 (201040_at), score: 0.61 GNB2guanine nucleotide binding protein (G protein), beta polypeptide 2 (200852_x_at), score: 0.53 GP1BBglycoprotein Ib (platelet), beta polypeptide (206655_s_at), score: 0.59 GPR176G protein-coupled receptor 176 (206673_at), score: 0.52 GPS1G protein pathway suppressor 1 (217782_s_at), score: 0.52 GRINAglutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) (212090_at), score: 0.7 GTPBP1GTP binding protein 1 (219357_at), score: 0.58 GZMMgranzyme M (lymphocyte met-ase 1) (207460_at), score: -0.64 H1FXH1 histone family, member X (204805_s_at), score: 0.54 HAB1B1 for mucin (215778_x_at), score: -0.83 HAMPhepcidin antimicrobial peptide (220491_at), score: -0.69 HAND1heart and neural crest derivatives expressed 1 (220138_at), score: -0.71 HDAC5histone deacetylase 5 (202455_at), score: 0.68 HIST1H4Dhistone cluster 1, H4d (208076_at), score: -0.63 HLA-DQB1major histocompatibility complex, class II, DQ beta 1 (211654_x_at), score: -0.63 HLA-Emajor histocompatibility complex, class I, E (200904_at), score: 0.57 HMG20Bhigh-mobility group 20B (210719_s_at), score: 0.52 HMGA1high mobility group AT-hook 1 (210457_x_at), score: 0.78 HNRNPUL1heterogeneous nuclear ribonucleoprotein U-like 1 (209675_s_at), score: 0.56 HS3ST2heparan sulfate (glucosamine) 3-O-sulfotransferase 2 (219697_at), score: -0.62 HSD17B6hydroxysteroid (17-beta) dehydrogenase 6 homolog (mouse) (37512_at), score: -0.74 HSF1heat shock transcription factor 1 (202344_at), score: 0.79 IDUAiduronidase, alpha-L- (205059_s_at), score: 0.76 IGFBP4insulin-like growth factor binding protein 4 (201508_at), score: 0.58 IGHMBP2immunoglobulin mu binding protein 2 (215980_s_at), score: -0.62 IL22RA1interleukin 22 receptor, alpha 1 (220056_at), score: -0.7 INF2inverted formin, FH2 and WH2 domain containing (218144_s_at), score: 0.55 INTS1integrator complex subunit 1 (212212_s_at), score: 0.68 ISYNA1inositol-3-phosphate synthase 1 (222240_s_at), score: 0.52 ITGA3integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) (201474_s_at), score: 0.56 JMJD7jumonji domain containing 7 (60528_at), score: -0.64 JUNDjun D proto-oncogene (203751_x_at), score: 0.74 JUPjunction plakoglobin (201015_s_at), score: 0.86 KAL1Kallmann syndrome 1 sequence (205206_at), score: -0.72 KCTD13potassium channel tetramerisation domain containing 13 (45653_at), score: 0.53 KCTD3potassium channel tetramerisation domain containing 3 (217894_at), score: -0.63 KDELR1KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1 (200922_at), score: 0.66 KIAA0892KIAA0892 (212505_s_at), score: 0.56 KIAA1009KIAA1009 (206005_s_at), score: -0.65 KIAA1305KIAA1305 (220911_s_at), score: 0.59 KIAA1539KIAA1539 (207765_s_at), score: 0.59 KLHL23kelch-like 23 (Drosophila) (213610_s_at), score: -0.71 KLHL26kelch-like 26 (Drosophila) (219354_at), score: 0.67 KRT9keratin 9 (208188_at), score: -0.63 LEPREL2leprecan-like 2 (204854_at), score: 0.75 LIMS2LIM and senescent cell antigen-like domains 2 (220765_s_at), score: 0.71 LMF2lipase maturation factor 2 (212682_s_at), score: 0.64 LOC284440hypothetical LOC284440 (216126_at), score: -0.69 LOC440434hypothetical protein FLJ11822 (215090_x_at), score: 0.66 LPGAT1lysophosphatidylglycerol acyltransferase 1 (202651_at), score: -0.66 LPPR2lipid phosphate phosphatase-related protein type 2 (218509_at), score: 0.54 LRDDleucine-rich repeats and death domain containing (221640_s_at), score: 0.55 LRP5low density lipoprotein receptor-related protein 5 (209468_at), score: 0.57 LRRC41leucine rich repeat containing 41 (201932_at), score: 0.58 LTBP3latent transforming growth factor beta binding protein 3 (219922_s_at), score: 0.6 LTBRlymphotoxin beta receptor (TNFR superfamily, member 3) (203005_at), score: 0.55 LY6Hlymphocyte antigen 6 complex, locus H (206773_at), score: -0.7 MAGOH2mago-nashi homolog 2, proliferation-associated (Drosophila) (217693_x_at), score: -0.86 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: 0.92 MAP2K2mitogen-activated protein kinase kinase 2 (213490_s_at), score: 0.74 MAP3K11mitogen-activated protein kinase kinase kinase 11 (203652_at), score: 0.62 MAP3K2mitogen-activated protein kinase kinase kinase 2 (221695_s_at), score: 0.58 MAP3K6mitogen-activated protein kinase kinase kinase 6 (219278_at), score: 0.75 MAP4microtubule-associated protein 4 (200836_s_at), score: 0.69 MAP7D1MAP7 domain containing 1 (217943_s_at), score: 0.68 MAPK3mitogen-activated protein kinase 3 (212046_x_at), score: 0.56 MAPKAPK2mitogen-activated protein kinase-activated protein kinase 2 (201461_s_at), score: 0.54 MAPKAPK5mitogen-activated protein kinase-activated protein kinase 5 (212871_at), score: 0.54 MAZMYC-associated zinc finger protein (purine-binding transcription factor) (207824_s_at), score: 0.52 MBD3methyl-CpG binding domain protein 3 (41160_at), score: 0.6 MBOAT7membrane bound O-acyltransferase domain containing 7 (209179_s_at), score: 0.61 MGAT1mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase (201126_s_at), score: 0.67 MGAT4Bmannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme B (220189_s_at), score: 0.71 MKL1megakaryoblastic leukemia (translocation) 1 (212748_at), score: 0.59 MN1meningioma (disrupted in balanced translocation) 1 (205330_at), score: 0.52 MRPS2mitochondrial ribosomal protein S2 (218001_at), score: 0.6 MUC1mucin 1, cell surface associated (207847_s_at), score: 0.72 MUC5ACmucin 5AC, oligomeric mucus/gel-forming (217182_at), score: -0.66 MUL1mitochondrial E3 ubiquitin ligase 1 (218246_at), score: 0.53 MYL4myosin, light chain 4, alkali; atrial, embryonic (210088_x_at), score: -0.73 MYO15Amyosin XVA (220288_at), score: -0.73 MYO9Bmyosin IXB (217297_s_at), score: 0.67 NACAP1nascent-polypeptide-associated complex alpha polypeptide pseudogene 1 (211445_x_at), score: -0.7 NARG1LNMDA receptor regulated 1-like (219378_at), score: -0.72 NCDNneurochondrin (209556_at), score: 0.71 NCKAP1NCK-associated protein 1 (217465_at), score: -0.93 NCLNnicalin homolog (zebrafish) (222206_s_at), score: 0.54 NCOR2nuclear receptor co-repressor 2 (207760_s_at), score: 0.55 NCRNA00093non-protein coding RNA 93 (210723_x_at), score: -0.69 NFATC4nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 (205897_at), score: 0.56 NFKB1nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 (209239_at), score: 0.54 NFKB2nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) (207535_s_at), score: 0.54 NID1nidogen 1 (202008_s_at), score: 0.58 NINJ1ninjurin 1 (203045_at), score: 0.54 NOL10nucleolar protein 10 (218591_s_at), score: -0.67 NOTCH2Notch homolog 2 (Drosophila) (210756_s_at), score: 0.54 NOTCH3Notch homolog 3 (Drosophila) (203238_s_at), score: 0.58 NOVA2neuro-oncological ventral antigen 2 (206477_s_at), score: -0.74 NPDC1neural proliferation, differentiation and control, 1 (218086_at), score: 0.78 NXT2nuclear transport factor 2-like export factor 2 (209628_at), score: -0.68 OBFC2Boligonucleotide/oligosaccharide-binding fold containing 2B (218903_s_at), score: 0.58 OGDHoxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) (201282_at), score: 0.66 OR7E47Polfactory receptor, family 7, subfamily E, member 47 pseudogene (216698_x_at), score: -0.76 PARVBparvin, beta (204629_at), score: 0.83 PCDHG@protocadherin gamma cluster (215836_s_at), score: 0.77 PCDHGA1protocadherin gamma subfamily A, 1 (209079_x_at), score: 0.69 PCDHGA11protocadherin gamma subfamily A, 11 (211876_x_at), score: 0.72 PCDHGA3protocadherin gamma subfamily A, 3 (216352_x_at), score: 0.69 PCTK1PCTAIRE protein kinase 1 (208824_x_at), score: 0.53 PDE7Bphosphodiesterase 7B (220343_at), score: -0.65 PDLIM7PDZ and LIM domain 7 (enigma) (203370_s_at), score: 0.57 PERLD1per1-like domain containing 1 (55616_at), score: -0.66 PHF7PHD finger protein 7 (215622_x_at), score: -0.79 PHLDA3pleckstrin homology-like domain, family A, member 3 (218634_at), score: 0.64 PI4K2Aphosphatidylinositol 4-kinase type 2 alpha (209345_s_at), score: 0.54 PIGOphosphatidylinositol glycan anchor biosynthesis, class O (209998_at), score: 0.66 PIK3R2phosphoinositide-3-kinase, regulatory subunit 2 (beta) (207105_s_at), score: 0.66 PINK1PTEN induced putative kinase 1 (209018_s_at), score: 0.58 PIP5K1Cphosphatidylinositol-4-phosphate 5-kinase, type I, gamma (212518_at), score: 0.93 PKN1protein kinase N1 (202161_at), score: 0.79 PLAUplasminogen activator, urokinase (211668_s_at), score: 0.66 PLD3phospholipase D family, member 3 (201050_at), score: 0.66 PLEC1plectin 1, intermediate filament binding protein 500kDa (216971_s_at), score: 0.57 PLEKHM2pleckstrin homology domain containing, family M (with RUN domain) member 2 (212146_at), score: 0.58 PLK3polo-like kinase 3 (Drosophila) (204958_at), score: 0.54 PLSCR3phospholipid scramblase 3 (218828_at), score: 0.6 PLXNB1plexin B1 (215807_s_at), score: 0.65 PLXNB2plexin B2 (208890_s_at), score: 0.57 PNPLA6patatin-like phospholipase domain containing 6 (203718_at), score: 0.76 POLR2Apolymerase (RNA) II (DNA directed) polypeptide A, 220kDa (202725_at), score: 0.66 POLRMTpolymerase (RNA) mitochondrial (DNA directed) (203782_s_at), score: 0.64 POM121POM121 membrane glycoprotein (rat) (212178_s_at), score: 0.67 POMZP3POM (POM121 homolog, rat) and ZP3 fusion (204148_s_at), score: 0.52 PORP450 (cytochrome) oxidoreductase (208928_at), score: 0.64 PPIGpeptidylprolyl isomerase G (cyclophilin G) (208993_s_at), score: -0.66 PPP2R5Bprotein phosphatase 2, regulatory subunit B', beta isoform (635_s_at), score: 0.57 PRKACAprotein kinase, cAMP-dependent, catalytic, alpha (202801_at), score: 0.68 PRKCDprotein kinase C, delta (202545_at), score: 0.74 PRKD2protein kinase D2 (209282_at), score: 0.53 PTOV1prostate tumor overexpressed 1 (212032_s_at), score: 0.66 PTPN14protein tyrosine phosphatase, non-receptor type 14 (205503_at), score: 0.7 PTPROprotein tyrosine phosphatase, receptor type, O (211600_at), score: 0.62 PXNpaxillin (211823_s_at), score: 0.79 PYGBphosphorylase, glycogen; brain (201481_s_at), score: 0.59 RAB11BRAB11B, member RAS oncogene family (34478_at), score: 0.76 RAB11FIP3RAB11 family interacting protein 3 (class II) (203933_at), score: 0.6 RANBP3RAN binding protein 3 (210120_s_at), score: 0.77 RASA3RAS p21 protein activator 3 (206220_s_at), score: 0.56 RASA4RAS p21 protein activator 4 (208534_s_at), score: 0.52 RBM15BRNA binding motif protein 15B (202689_at), score: 0.62 RBM42RNA binding motif protein 42 (205740_s_at), score: 0.55 RELAv-rel reticuloendotheliosis viral oncogene homolog A (avian) (209878_s_at), score: 0.59 REREarginine-glutamic acid dipeptide (RE) repeats (200940_s_at), score: 0.54 RFNGRFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase (212968_at), score: 0.54 RHOGras homolog gene family, member G (rho G) (203175_at), score: 0.56 RIPK1receptor (TNFRSF)-interacting serine-threonine kinase 1 (209941_at), score: 0.79 RLN1relaxin 1 (211753_s_at), score: -0.78 RNF11ring finger protein 11 (208924_at), score: -0.64 RNF123ring finger protein 123 (221063_x_at), score: 0.53 RNF126ring finger protein 126 (205748_s_at), score: 0.62 RNF130ring finger protein 130 (217865_at), score: 0.57 RNF24ring finger protein 24 (204669_s_at), score: 0.55 RNF40ring finger protein 40 (206845_s_at), score: 0.63 RP3-377H14.5hypothetical LOC285830 (222279_at), score: -0.71 RP5-1000E10.4suppressor of IKK epsilon (221705_s_at), score: -0.77 RP5-886K2.1neuronal thread protein AD7c-NTP (207953_at), score: -0.7 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: -0.64 RPL21P68ribosomal protein L21 pseudogene 68 (217340_at), score: -0.72 RPL38ribosomal protein L38 (202028_s_at), score: -0.63 RPRD2regulation of nuclear pre-mRNA domain containing 2 (212553_at), score: 0.53 RPS17P5ribosomal protein S17 pseudogene 5 (216348_at), score: -0.83 RPS25ribosomal protein S25 (200091_s_at), score: -0.62 S100A14S100 calcium binding protein A14 (218677_at), score: -0.7 SAV1salvador homolog 1 (Drosophila) (218276_s_at), score: 0.6 SBF1SET binding factor 1 (39835_at), score: 0.74 SBNO2strawberry notch homolog 2 (Drosophila) (204166_at), score: 0.81 SCAMP4secretory carrier membrane protein 4 (213244_at), score: 0.85 SCAMP5secretory carrier membrane protein 5 (212699_at), score: -0.69 SCAPSREBF chaperone (212329_at), score: 0.56 SDAD1SDA1 domain containing 1 (218607_s_at), score: 0.74 SDC3syndecan 3 (202898_at), score: 0.69 SDPRserum deprivation response (phosphatidylserine binding protein) (218711_s_at), score: 0.56 SENP3SUMO1/sentrin/SMT3 specific peptidase 3 (215113_s_at), score: 0.61 SFNstratifin (33322_i_at), score: -0.67 SFRP1secreted frizzled-related protein 1 (202036_s_at), score: 0.53 SH3GLB2SH3-domain GRB2-like endophilin B2 (218813_s_at), score: 0.73 SHBSrc homology 2 domain containing adaptor protein B (204657_s_at), score: 0.68 SIGIRRsingle immunoglobulin and toll-interleukin 1 receptor (TIR) domain (52940_at), score: 0.77 SIGLEC1sialic acid binding Ig-like lectin 1, sialoadhesin (44673_at), score: -0.63 SIGLEC6sialic acid binding Ig-like lectin 6 (210796_x_at), score: -0.79 SIN3BSIN3 homolog B, transcription regulator (yeast) (209352_s_at), score: 0.6 SLAMF1signaling lymphocytic activation molecule family member 1 (206181_at), score: -0.63 SLC1A7solute carrier family 1 (glutamate transporter), member 7 (210923_at), score: -0.63 SLC25A1solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1 (210010_s_at), score: 0.56 SLC2A8solute carrier family 2 (facilitated glucose transporter), member 8 (218985_at), score: 0.64 SLC35C1solute carrier family 35, member C1 (218485_s_at), score: 0.61 SLC4A2solute carrier family 4, anion exchanger, member 2 (erythrocyte membrane protein band 3-like 1) (202111_at), score: 0.58 SLC5A5solute carrier family 5 (sodium iodide symporter), member 5 (211123_at), score: -0.76 SLC9A1solute carrier family 9 (sodium/hydrogen exchanger), member 1 (209453_at), score: 0.69 SLC9A3R2solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 (209830_s_at), score: 0.77 SMAD7SMAD family member 7 (204790_at), score: 0.53 SMG6Smg-6 homolog, nonsense mediated mRNA decay factor (C. elegans) (214940_s_at), score: 0.81 SMN1survival of motor neuron 1, telomeric (203852_s_at), score: -0.64 SMTNsmoothelin (209427_at), score: 0.64 SMYD5SMYD family member 5 (209516_at), score: 0.57 SNAPC2small nuclear RNA activating complex, polypeptide 2, 45kDa (204104_at), score: 0.64 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: 0.76 SORBS3sorbin and SH3 domain containing 3 (209253_at), score: 0.58 SP3Sp3 transcription factor (213168_at), score: -0.63 SPAG8sperm associated antigen 8 (206816_s_at), score: -0.69 SPARCL1SPARC-like 1 (hevin) (200795_at), score: -0.64 SPATA20spermatogenesis associated 20 (218164_at), score: 0.57 SPG7spastic paraplegia 7 (pure and complicated autosomal recessive) (202104_s_at), score: 0.6 SPRY4sprouty homolog 4 (Drosophila) (221489_s_at), score: 0.61 SSBP3single stranded DNA binding protein 3 (217991_x_at), score: 0.87 SSX3synovial sarcoma, X breakpoint 3 (211731_x_at), score: -0.63 ST6GALNAC4ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 (221551_x_at), score: 0.54 STRN4striatin, calmodulin binding protein 4 (217903_at), score: 0.63 SULT1B1sulfotransferase family, cytosolic, 1B, member 1 (207601_at), score: -0.79 SUPT6Hsuppressor of Ty 6 homolog (S. cerevisiae) (208831_x_at), score: 0.67 SYNPOsynaptopodin (202796_at), score: 0.6 TAF15TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa (202840_at), score: -0.65 TAF4BTAF4b RNA polymerase II, TATA box binding protein (TBP)-associated factor, 105kDa (216226_at), score: -0.63 TAL1T-cell acute lymphocytic leukemia 1 (206283_s_at), score: -0.62 TAOK2TAO kinase 2 (204986_s_at), score: 0.58 TAPBPTAP binding protein (tapasin) (208829_at), score: 0.7 TAS2R14taste receptor, type 2, member 14 (221391_at), score: -0.68 TENC1tensin like C1 domain containing phosphatase (tensin 2) (212494_at), score: 0.57 TERF2telomeric repeat binding factor 2 (203611_at), score: 0.59 TGFB1transforming growth factor, beta 1 (203085_s_at), score: 0.64 TGFBR2transforming growth factor, beta receptor II (70/80kDa) (207334_s_at), score: 0.61 TGM2transglutaminase 2 (C polypeptide, protein-glutamine-gamma-glutamyltransferase) (201042_at), score: 0.53 TGM4transglutaminase 4 (prostate) (217566_s_at), score: -0.65 THUMPD1THUMP domain containing 1 (206555_s_at), score: -0.64 TIAF1TGFB1-induced anti-apoptotic factor 1 (202039_at), score: 0.63 TIMP2TIMP metallopeptidase inhibitor 2 (203167_at), score: 0.54 TLE3transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) (206472_s_at), score: 0.61 TMCC2transmembrane and coiled-coil domain family 2 (213096_at), score: -0.69 TMEM184Ctransmembrane protein 184C (219074_at), score: -0.77 TMEM222transmembrane protein 222 (221512_at), score: 0.57 TMEM8transmembrane protein 8 (five membrane-spanning domains) (221882_s_at), score: 0.58 TMSB4Ythymosin beta 4, Y-linked (206769_at), score: -0.77 TNFRSF8tumor necrosis factor receptor superfamily, member 8 (206729_at), score: -0.81 TOB2transducer of ERBB2, 2 (221496_s_at), score: 0.53 TOMM34translocase of outer mitochondrial membrane 34 (201870_at), score: 0.54 TRIM3tripartite motif-containing 3 (204911_s_at), score: 0.55 TSC2tuberous sclerosis 2 (215735_s_at), score: 0.76 TSKUtsukushin (218245_at), score: 0.72 TSSC4tumor suppressing subtransferable candidate 4 (218612_s_at), score: 0.58 TTC15tetratricopeptide repeat domain 15 (203122_at), score: 0.57 TUBA4Btubulin, alpha 4b (pseudogene) (207490_at), score: -0.85 TULP2tubby like protein 2 (206733_at), score: -0.71 TXLNAtaxilin alpha (212300_at), score: 0.6 TXNthioredoxin (216609_at), score: -0.74 UBA2ubiquitin-like modifier activating enzyme 2 (201177_s_at), score: -0.64 UBAC1UBA domain containing 1 (202151_s_at), score: 0.58 UBTD1ubiquitin domain containing 1 (219172_at), score: 0.79 UBXN6UBX domain protein 6 (220757_s_at), score: 0.59 USF2upstream transcription factor 2, c-fos interacting (202152_x_at), score: 0.72 VAC14Vac14 homolog (S. cerevisiae) (218169_at), score: 0.56 VAT1vesicle amine transport protein 1 homolog (T. californica) (208626_s_at), score: 0.65 VCPIP1valosin containing protein (p97)/p47 complex interacting protein 1 (219810_at), score: -0.67 VEGFBvascular endothelial growth factor B (203683_s_at), score: 1 VPS37Bvacuolar protein sorting 37 homolog B (S. cerevisiae) (221704_s_at), score: 0.83 WBP2WW domain binding protein 2 (209117_at), score: 0.65 WDR4WD repeat domain 4 (221632_s_at), score: -0.63 WDR42AWD repeat domain 42A (202249_s_at), score: 0.68 WHSC1L1Wolf-Hirschhorn syndrome candidate 1-like 1 (218173_s_at), score: -0.7 WNT6wingless-type MMTV integration site family, member 6 (71933_at), score: -0.65 XYLBxylulokinase homolog (H. influenzae) (214776_x_at), score: -0.7 ZBTB24zinc finger and BTB domain containing 24 (205340_at), score: -0.65 ZC3H3zinc finger CCCH-type containing 3 (213445_at), score: 0.55 ZDHHC14zinc finger, DHHC-type containing 14 (219247_s_at), score: -0.71 ZER1zer-1 homolog (C. elegans) (202456_s_at), score: 0.53 ZFHX3zinc finger homeobox 3 (208033_s_at), score: 0.83 ZFP36L1zinc finger protein 36, C3H type-like 1 (211965_at), score: 0.53 ZNF107zinc finger protein 107 (205739_x_at), score: -0.7 ZNF248zinc finger protein 248 (213269_at), score: -0.66 ZNF282zinc finger protein 282 (212892_at), score: 0.71 ZNF574zinc finger protein 574 (218762_at), score: 0.53 ZNF580zinc finger protein 580 (220748_s_at), score: 0.57 ZPBPzona pellucida binding protein (207021_at), score: -0.71 ZYXzyxin (200808_s_at), score: 0.62
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-3407-raw-cel-1437949557.cel | 1 | 4 | Cockayne | hgu133a | CS | eGFP |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3407-raw-cel-1437949655.cel | 3 | 4 | Cockayne | hgu133a | none | CSB |
| 46C.CEL | 3 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 3 |
| E-GEOD-3860-raw-cel-1561690199.cel | 1 | 5 | HGPS | hgu133a | none | GM0316B |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 10358_WBS.CEL | 1 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| E-TABM-263-raw-cel-1515485991.cel | 18 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 46A.CEL | 1 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 1 |
| E-TABM-263-raw-cel-1515486071.cel | 22 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486411.cel | 39 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |