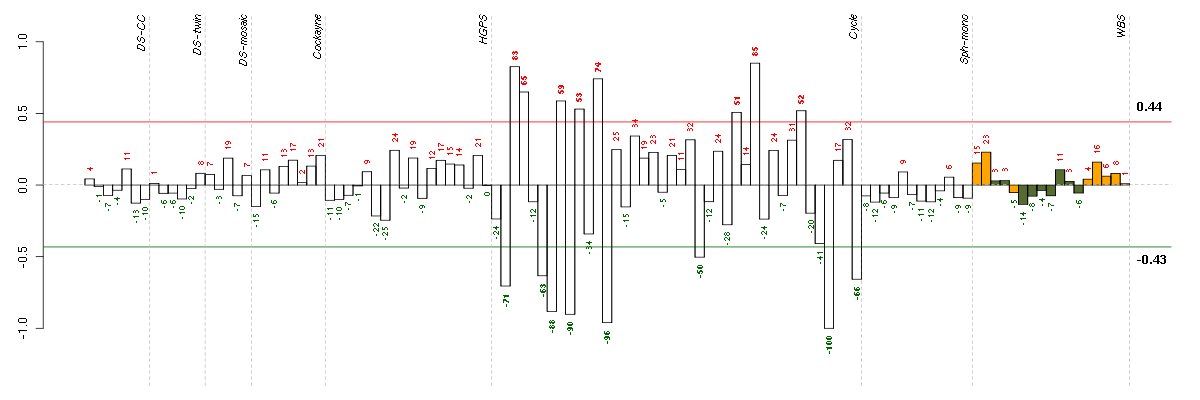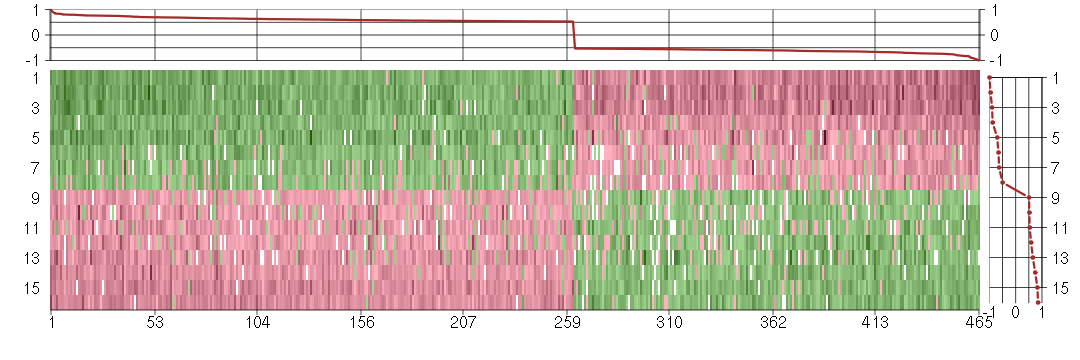

Under-expression is coded with green,
over-expression with red color.

MAPKKK cascade
Cascade of at least three protein kinase activities culminating in the phosphorylation and activation of a MAP kinase. MAPKKK cascades lie downstream of numerous signaling pathways.
activation of MAPK activity
The initiation of the activity of the inactive enzyme MAP kinase by phosphorylation by a MAPKK.
metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
signal transduction
The cascade of processes by which a signal interacts with a receptor, causing a change in the level or activity of a second messenger or other downstream target, and ultimately effecting a change in the functioning of the cell.
protein modification process
The covalent alteration of one or more amino acids occurring in proteins, peptides and nascent polypeptides (co-translational, post-translational modifications). Includes the modification of charged tRNAs that are destined to occur in a protein (pre-translation modification).
protein amino acid phosphorylation
The process of introducing a phosphate group on to a protein.
phosphorus metabolic process
The chemical reactions and pathways involving the nonmetallic element phosphorus or compounds that contain phosphorus, usually in the form of a phosphate group (PO4).
phosphate metabolic process
The chemical reactions and pathways involving the phosphate group, the anion or salt of any phosphoric acid.
cell communication
Any process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
intracellular signaling cascade
A series of reactions within the cell that occur as a result of a single trigger reaction or compound.
protein kinase cascade
A series of reactions, mediated by protein kinases, which occurs as a result of a single trigger reaction or compound.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
phosphorylation
The process of introducing a phosphate group into a molecule, usually with the formation of a phosphoric ester, a phosphoric anhydride or a phosphoric amide.
protein metabolic process
The chemical reactions and pathways involving a specific protein, rather than of proteins in general. Includes protein modification.
positive regulation of kinase activity
Any process that activates or increases the frequency, rate or extent of kinase activity, the catalysis of the transfer of a phosphate group, usually from ATP, to a substrate molecule.
cellular biopolymer metabolic process
The chemical reactions and pathways involving biopolymers, long, repeating chains of monomers found in nature, such as polysaccharides and proteins, as carried out by individual cells.
positive regulation of catalytic activity
Any process that activates or increases the activity of an enzyme.
macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
biopolymer metabolic process
The chemical reactions and pathways involving biopolymers, long, repeating chains of monomers found in nature, such as polysaccharides and proteins.
regulation of MAP kinase activity
Any process that modulates the frequency, rate or extent of MAP kinase activity.
positive regulation of MAP kinase activity
Any process that activates or increases the frequency, rate or extent of MAP kinase activity.
biopolymer modification
The covalent alteration of one or more monomeric units in a polypeptide, polynucleotide, polysaccharide, or other biological polymer, resulting in a change in its properties.
regulation of JUN kinase activity
Any process that modulates the frequency, rate or extent of JUN kinase activity.
positive regulation of JUN kinase activity
Any process that activates or increases the frequency, rate or extent of JUN kinase activity.
regulation of kinase activity
Any process that modulates the frequency, rate or extent of kinase activity, the catalysis of the transfer of a phosphate group, usually from ATP, to a substrate molecule.
post-translational protein modification
The covalent alteration of one or more amino acids occurring in a protein after the protein has been completely translated and released from the ribosome.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
primary metabolic process
The chemical reactions and pathways involving those compounds which are formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
cellular protein metabolic process
The chemical reactions and pathways involving a specific protein, rather than of proteins in general, occurring at the level of an individual cell. Includes protein modification.
regulation of protein kinase activity
Any process that modulates the frequency, rate or extent of protein kinase activity.
positive regulation of protein kinase activity
Any process that activates or increases the frequency, rate or extent of protein kinase activity.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of catalytic activity
Any process that modulates the activity of an enzyme.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of transferase activity
Any process that modulates the frequency, rate or extent of transferase activity, the catalysis of the transfer of a group, e.g. a methyl group, glycosyl group, acyl group, phosphorus-containing, or other groups, from one compound (generally regarded as the donor) to another compound (generally regarded as the acceptor). Transferase is the systematic name for any enzyme of EC class 2.
positive regulation of transferase activity
Any process that activates or increases the frequency, rate or extent of transferase activity, the catalysis of the transfer of a group, e.g. a methyl group, glycosyl group, acyl group, phosphorus-containing, or other groups, from a donor compound to an acceptor.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
regulation of molecular function
Any process that modulates the frequency, rate or extent of molecular functions. Molecular functions are elemental biological activities occurring at the molecular level, such as catalysis or binding.
all
This term is the most general term possible
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
signal transduction
The cascade of processes by which a signal interacts with a receptor, causing a change in the level or activity of a second messenger or other downstream target, and ultimately effecting a change in the functioning of the cell.
protein metabolic process
The chemical reactions and pathways involving a specific protein, rather than of proteins in general. Includes protein modification.
cellular biopolymer metabolic process
The chemical reactions and pathways involving biopolymers, long, repeating chains of monomers found in nature, such as polysaccharides and proteins, as carried out by individual cells.
cellular protein metabolic process
The chemical reactions and pathways involving a specific protein, rather than of proteins in general, occurring at the level of an individual cell. Includes protein modification.
protein modification process
The covalent alteration of one or more amino acids occurring in proteins, peptides and nascent polypeptides (co-translational, post-translational modifications). Includes the modification of charged tRNAs that are destined to occur in a protein (pre-translation modification).
positive regulation of transferase activity
Any process that activates or increases the frequency, rate or extent of transferase activity, the catalysis of the transfer of a group, e.g. a methyl group, glycosyl group, acyl group, phosphorus-containing, or other groups, from a donor compound to an acceptor.
positive regulation of kinase activity
Any process that activates or increases the frequency, rate or extent of kinase activity, the catalysis of the transfer of a phosphate group, usually from ATP, to a substrate molecule.
protein amino acid phosphorylation
The process of introducing a phosphate group on to a protein.
activation of MAPK activity
The initiation of the activity of the inactive enzyme MAP kinase by phosphorylation by a MAPKK.
positive regulation of protein kinase activity
Any process that activates or increases the frequency, rate or extent of protein kinase activity.
positive regulation of MAP kinase activity
Any process that activates or increases the frequency, rate or extent of MAP kinase activity.
activation of MAPK activity
The initiation of the activity of the inactive enzyme MAP kinase by phosphorylation by a MAPKK.
positive regulation of JUN kinase activity
Any process that activates or increases the frequency, rate or extent of JUN kinase activity.

intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
intermediate filament
A cytoskeletal structure that forms a distinct elongated structure, characteristically 10 nm in diameter, that occurs in the cytoplasm of eukaryotic cells. Intermediate filaments form a fibrous system, composed of chemically heterogeneous subunits and involved in mechanically integrating the various components of the cytoplasmic space. Intermediate filaments may be divided into five chemically distinct classes: Type I, acidic keratins; Type II, basic keratins; Type III, including desmin, vimentin and others; Type IV, neurofilaments and related filaments; and Type V, lamins.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
cytoskeleton
Any of the various filamentous elements that form the internal framework of cells, and typically remain after treatment of the cells with mild detergent to remove membrane constituents and soluble components of the cytoplasm. The term embraces intermediate filaments, microfilaments, microtubules, the microtrabecular lattice, and other structures characterized by a polymeric filamentous nature and long-range order within the cell. The various elements of the cytoskeleton not only serve in the maintenance of cellular shape but also have roles in other cellular functions, including cellular movement, cell division, endocytosis, and movement of organelles.
neurofilament
A type of intermediate filament found in the core of neuronal axons. Neurofilaments are heteropolymers composed of three type IV polypeptides: NF-L, NF-M, and NF-H (for low, middle, and high molecular weight). Neurofilaments are responsible for the radial growth of an axon and determine axonal diameter.
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane. Includes ribosomes, the cytoskeleton and chromosomes.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane and occurring within the cell. Includes ribosomes, the cytoskeleton and chromosomes.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
cytoskeletal part
Any constituent part of the cytoskeleton, a cellular scaffolding or skeleton that maintains cell shape, enables some cell motion (using structures such as flagella and cilia), and plays important roles in both intra-cellular transport (e.g. the movement of vesicles and organelles) and cellular division. Includes constituent parts of intermediate filaments, microfilaments, microtubules, and the microtrabecular lattice.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
intermediate filament cytoskeleton
Cytoskeletal structure made from intermediate filaments, typically organized in the cytosol as an extended system that stretches from the nuclear envelope to the plasma membrane. Some intermediate filaments run parallel to the cell surface, while others traverse the cytosol; together they form an internal framework that helps support the shape and resilience of the cell.
neurofilament cytoskeleton
Intermediate filament cytoskeletal structure that is made up of neurofilaments. Neurofilaments are specialized intermediate filaments found in neurons.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
intracellular non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane and occurring within the cell. Includes ribosomes, the cytoskeleton and chromosomes.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
cytoskeletal part
Any constituent part of the cytoskeleton, a cellular scaffolding or skeleton that maintains cell shape, enables some cell motion (using structures such as flagella and cilia), and plays important roles in both intra-cellular transport (e.g. the movement of vesicles and organelles) and cellular division. Includes constituent parts of intermediate filaments, microfilaments, microtubules, and the microtrabecular lattice.
intermediate filament
A cytoskeletal structure that forms a distinct elongated structure, characteristically 10 nm in diameter, that occurs in the cytoplasm of eukaryotic cells. Intermediate filaments form a fibrous system, composed of chemically heterogeneous subunits and involved in mechanically integrating the various components of the cytoplasmic space. Intermediate filaments may be divided into five chemically distinct classes: Type I, acidic keratins; Type II, basic keratins; Type III, including desmin, vimentin and others; Type IV, neurofilaments and related filaments; and Type V, lamins.
neurofilament
A type of intermediate filament found in the core of neuronal axons. Neurofilaments are heteropolymers composed of three type IV polypeptides: NF-L, NF-M, and NF-H (for low, middle, and high molecular weight). Neurofilaments are responsible for the radial growth of an axon and determine axonal diameter.

AAASachalasia, adrenocortical insufficiency, alacrimia (Allgrove, triple-A) (218075_at), score: 0.63 ABCA7ATP-binding cassette, sub-family A (ABC1), member 7 (219577_s_at), score: -0.64 ABCA8ATP-binding cassette, sub-family A (ABC1), member 8 (204719_at), score: 0.58 ABCD1ATP-binding cassette, sub-family D (ALD), member 1 (205142_x_at), score: 0.67 ABLIM1actin binding LIM protein 1 (200965_s_at), score: 0.69 ACSL1acyl-CoA synthetase long-chain family member 1 (201963_at), score: -0.54 ACSL4acyl-CoA synthetase long-chain family member 4 (202422_s_at), score: -0.69 ACSS3acyl-CoA synthetase short-chain family member 3 (219616_at), score: 0.56 ACTN3actinin, alpha 3 (206891_at), score: -0.55 ACTR3BARP3 actin-related protein 3 homolog B (yeast) (218868_at), score: -0.75 ADAadenosine deaminase (204639_at), score: -0.63 ADAM19ADAM metallopeptidase domain 19 (meltrin beta) (209765_at), score: -0.54 ADARadenosine deaminase, RNA-specific (201786_s_at), score: 0.63 ADCK2aarF domain containing kinase 2 (221893_s_at), score: 0.57 ADCY3adenylate cyclase 3 (209321_s_at), score: 0.53 ADORA2Badenosine A2b receptor (205891_at), score: 0.69 AGPAT11-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) (215535_s_at), score: 0.66 AGTangiotensinogen (serpin peptidase inhibitor, clade A, member 8) (202834_at), score: -0.55 AHI1Abelson helper integration site 1 (221569_at), score: -0.58 AIM1absent in melanoma 1 (212543_at), score: 0.77 AKAP2A kinase (PRKA) anchor protein 2 (202759_s_at), score: 0.61 ALDH1L1aldehyde dehydrogenase 1 family, member L1 (215798_at), score: -0.74 ANXA10annexin A10 (210143_at), score: -0.61 APPamyloid beta (A4) precursor protein (214953_s_at), score: -0.54 ARFGAP3ADP-ribosylation factor GTPase activating protein 3 (202211_at), score: -0.54 ARHGDIARho GDP dissociation inhibitor (GDI) alpha (213606_s_at), score: 0.55 ARL6IP1ADP-ribosylation factor-like 6 interacting protein 1 (211935_at), score: 0.58 ATF5activating transcription factor 5 (204999_s_at), score: 0.6 ATMataxia telangiectasia mutated (210858_x_at), score: -0.58 ATN1atrophin 1 (40489_at), score: 0.55 ATP2C1ATPase, Ca++ transporting, type 2C, member 1 (211137_s_at), score: -0.71 ATP8B1ATPase, class I, type 8B, member 1 (214594_x_at), score: -0.57 B3GAT3beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) (203452_at), score: 0.54 B9D1B9 protein domain 1 (210534_s_at), score: 0.58 BAIAP2BAI1-associated protein 2 (209502_s_at), score: 0.58 BAP1BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) (201419_at), score: 0.62 BAT1HLA-B associated transcript 1 (200041_s_at), score: 0.59 BAT2HLA-B associated transcript 2 (212081_x_at), score: 0.75 BAT2LHLA-B associated transcript 2-like (212068_s_at), score: 0.56 BCL2L1BCL2-like 1 (215037_s_at), score: 0.65 BIRC3baculoviral IAP repeat-containing 3 (210538_s_at), score: 0.54 BMP6bone morphogenetic protein 6 (206176_at), score: -0.66 BTBD2BTB (POZ) domain containing 2 (207722_s_at), score: 0.69 C10orf97chromosome 10 open reading frame 97 (218297_at), score: -0.58 C11orf75chromosome 11 open reading frame 75 (219806_s_at), score: -0.64 C12orf29chromosome 12 open reading frame 29 (213701_at), score: -0.6 C14orf45chromosome 14 open reading frame 45 (220173_at), score: -0.68 C14orf94chromosome 14 open reading frame 94 (218383_at), score: 0.62 C16orf45chromosome 16 open reading frame 45 (212736_at), score: 0.53 C19orf61chromosome 19 open reading frame 61 (221335_x_at), score: 0.54 C1orf107chromosome 1 open reading frame 107 (214193_s_at), score: -0.53 C20orf117chromosome 20 open reading frame 117 (207711_at), score: 0.65 C20orf27chromosome 20 open reading frame 27 (50314_i_at), score: 0.57 C3orf64chromosome 3 open reading frame 64 (221935_s_at), score: -0.59 C4orf43chromosome 4 open reading frame 43 (218513_at), score: -0.73 C5orf44chromosome 5 open reading frame 44 (218674_at), score: -0.6 C7orf44chromosome 7 open reading frame 44 (209445_x_at), score: 0.53 C7orf58chromosome 7 open reading frame 58 (220032_at), score: 0.58 CABIN1calcineurin binding protein 1 (37652_at), score: 0.54 CABYRcalcium binding tyrosine-(Y)-phosphorylation regulated (219928_s_at), score: 0.59 CALB2calbindin 2 (205428_s_at), score: -0.82 CALCOCO1calcium binding and coiled-coil domain 1 (209002_s_at), score: 0.67 CAPN5calpain 5 (205166_at), score: 0.67 CASC3cancer susceptibility candidate 3 (207842_s_at), score: 0.71 CASP7caspase 7, apoptosis-related cysteine peptidase (207181_s_at), score: -0.58 CCDC101coiled-coil domain containing 101 (48117_at), score: -0.58 CCNFcyclin F (204826_at), score: 0.56 CD248CD248 molecule, endosialin (219025_at), score: 0.62 CDC25Bcell division cycle 25 homolog B (S. pombe) (201853_s_at), score: 0.7 CDC2L6cell division cycle 2-like 6 (CDK8-like) (212899_at), score: 0.66 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (217849_s_at), score: 0.75 CDC42EP1CDC42 effector protein (Rho GTPase binding) 1 (204693_at), score: 0.54 CDC42EP4CDC42 effector protein (Rho GTPase binding) 4 (214721_x_at), score: 0.8 CDCA3cell division cycle associated 3 (221436_s_at), score: 0.57 CDH13cadherin 13, H-cadherin (heart) (204726_at), score: -0.65 CDH6cadherin 6, type 2, K-cadherin (fetal kidney) (205532_s_at), score: -0.62 CDKN3cyclin-dependent kinase inhibitor 3 (209714_s_at), score: 0.6 CHEK2CHK2 checkpoint homolog (S. pombe) (210416_s_at), score: 0.53 CHMP5chromatin modifying protein 5 (218085_at), score: -0.58 CHRNB2cholinergic receptor, nicotinic, beta 2 (neuronal) (206635_at), score: -0.56 CHUKconserved helix-loop-helix ubiquitous kinase (209666_s_at), score: -0.64 CICcapicua homolog (Drosophila) (212784_at), score: 0.75 CITcitron (rho-interacting, serine/threonine kinase 21) (212801_at), score: 0.76 CIZ1CDKN1A interacting zinc finger protein 1 (205516_x_at), score: 0.66 CLN5ceroid-lipofuscinosis, neuronal 5 (204084_s_at), score: -0.59 CMPK1cytidine monophosphate (UMP-CMP) kinase 1, cytosolic (217870_s_at), score: -0.57 CNIH3cornichon homolog 3 (Drosophila) (214841_at), score: 0.63 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: 0.78 COL18A1collagen, type XVIII, alpha 1 (209081_s_at), score: 0.59 CORINcorin, serine peptidase (220356_at), score: -0.55 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: 0.8 CScitrate synthase (208660_at), score: 0.73 CSGALNACT2chondroitin sulfate N-acetylgalactosaminyltransferase 2 (222235_s_at), score: -0.58 CUX1cut-like homeobox 1 (214743_at), score: -0.6 DAXXdeath-domain associated protein (201763_s_at), score: 0.54 DCP2DCP2 decapping enzyme homolog (S. cerevisiae) (212919_at), score: -0.58 DDB2damage-specific DNA binding protein 2, 48kDa (203409_at), score: 0.63 DENND2ADENN/MADD domain containing 2A (53991_at), score: 0.62 DENND5BDENN/MADD domain containing 5B (215058_at), score: -0.64 DERL2Der1-like domain family, member 2 (218333_at), score: -0.59 DIS3DIS3 mitotic control homolog (S. cerevisiae) (218362_s_at), score: -0.57 DKFZP564O0823DKFZP564O0823 protein (204687_at), score: -0.69 DLSTPdihydrolipoamide S-succinyltransferase pseudogene (E2 component of 2-oxo-glutarate complex) (215210_s_at), score: -0.54 DMBT1deleted in malignant brain tumors 1 (208250_s_at), score: -0.56 DMPKdystrophia myotonica-protein kinase (37996_s_at), score: 0.73 DNAJB9DnaJ (Hsp40) homolog, subfamily B, member 9 (202843_at), score: -0.9 DNAJC6DnaJ (Hsp40) homolog, subfamily C, member 6 (204720_s_at), score: -0.72 DNM2dynamin 2 (202253_s_at), score: 0.55 DOCK9dedicator of cytokinesis 9 (212538_at), score: -0.54 DOPEY1dopey family member 1 (40612_at), score: -0.62 DSCR6Down syndrome critical region gene 6 (207267_s_at), score: 0.58 DSPPdentin sialophosphoprotein (221681_s_at), score: -0.56 DUS2Ldihydrouridine synthase 2-like, SMM1 homolog (S. cerevisiae) (47105_at), score: -0.55 DZIP3DAZ interacting protein 3, zinc finger (213186_at), score: 0.61 ECM1extracellular matrix protein 1 (209365_s_at), score: 0.6 EDEM1ER degradation enhancer, mannosidase alpha-like 1 (203279_at), score: -0.77 EDNRAendothelin receptor type A (204464_s_at), score: -0.54 EFHC2EF-hand domain (C-terminal) containing 2 (220591_s_at), score: 0.56 EGFepidermal growth factor (beta-urogastrone) (206254_at), score: -0.65 EHBP1L1EH domain binding protein 1-like 1 (221755_at), score: 0.55 EHMT2euchromatic histone-lysine N-methyltransferase 2 (202326_at), score: 0.56 EIF2AK3eukaryotic translation initiation factor 2-alpha kinase 3 (218696_at), score: -0.56 EIF4Eeukaryotic translation initiation factor 4E (201436_at), score: -0.54 EMCNendomucin (219436_s_at), score: 0.7 ENTPD1ectonucleoside triphosphate diphosphohydrolase 1 (209473_at), score: -0.65 EPHA4EPH receptor A4 (206114_at), score: -0.74 ERBB2IPerbb2 interacting protein (217941_s_at), score: -0.58 ESM1endothelial cell-specific molecule 1 (208394_x_at), score: -0.54 EVI1ecotropic viral integration site 1 (221884_at), score: -0.56 EVI2Becotropic viral integration site 2B (211742_s_at), score: 0.59 FABP3fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor) (214285_at), score: -0.54 FAM105Afamily with sequence similarity 105, member A (219694_at), score: 0.54 FAM18Bfamily with sequence similarity 18, member B (218446_s_at), score: -0.73 FAM62Afamily with sequence similarity 62 (C2 domain containing), member A (208858_s_at), score: 0.58 FAM64Afamily with sequence similarity 64, member A (221591_s_at), score: 0.53 FAT4FAT tumor suppressor homolog 4 (Drosophila) (219427_at), score: -0.59 FBXL14F-box and leucine-rich repeat protein 14 (213145_at), score: 0.8 FBXO17F-box protein 17 (220233_at), score: 0.58 FBXO2F-box protein 2 (219305_x_at), score: -0.56 FBXO31F-box protein 31 (219785_s_at), score: 0.54 FCF1FCF1 small subunit (SSU) processome component homolog (S. cerevisiae) (219927_at), score: -0.54 FGGYFGGY carbohydrate kinase domain containing (219718_at), score: 0.53 FICDFIC domain containing (219910_at), score: -0.64 FKBP10FK506 binding protein 10, 65 kDa (219249_s_at), score: -0.58 FLJ12529pre-mRNA cleavage factor I, 59 kDa subunit (217866_at), score: 0.64 FLOT1flotillin 1 (210142_x_at), score: 0.56 FNBP1formin binding protein 1 (212288_at), score: 0.56 FOLR3folate receptor 3 (gamma) (206371_at), score: -0.6 FOSL1FOS-like antigen 1 (204420_at), score: 0.61 FOSL2FOS-like antigen 2 (218880_at), score: 0.54 FOXC2forkhead box C2 (MFH-1, mesenchyme forkhead 1) (214520_at), score: 0.75 FOXF1forkhead box F1 (205935_at), score: 0.61 FOXK2forkhead box K2 (203064_s_at), score: 0.77 FOXM1forkhead box M1 (202580_x_at), score: 0.63 FRG1FSHD region gene 1 (204145_at), score: -0.64 FSTL3follistatin-like 3 (secreted glycoprotein) (203592_s_at), score: -0.58 FUBP3far upstream element (FUSE) binding protein 3 (212824_at), score: -0.53 G3BP2GTPase activating protein (SH3 domain) binding protein 2 (208841_s_at), score: -0.61 GABARAPL1GABA(A) receptor-associated protein like 1 (208868_s_at), score: -0.67 GABARAPL3GABA(A) receptors associated protein like 3 (pseudogene) (211458_s_at), score: -0.62 GFOD1glucose-fructose oxidoreductase domain containing 1 (219821_s_at), score: -0.83 GGA3golgi associated, gamma adaptin ear containing, ARF binding protein 3 (209411_s_at), score: 0.62 GHRgrowth hormone receptor (205498_at), score: -0.58 GKglycerol kinase (207387_s_at), score: -0.63 GK3Pglycerol kinase 3 pseudogene (215966_x_at), score: -0.81 GMCL1germ cell-less homolog 1 (Drosophila) (218458_at), score: -0.64 GMPRguanosine monophosphate reductase (204187_at), score: 0.72 GNL3Lguanine nucleotide binding protein-like 3 (nucleolar)-like (205010_at), score: -0.58 GOLGA8Ggolgi autoantigen, golgin subfamily a, 8G (222149_x_at), score: -0.59 GOLT1Bgolgi transport 1 homolog B (S. cerevisiae) (218193_s_at), score: -0.66 GPR56G protein-coupled receptor 56 (212070_at), score: 0.63 GPR65G protein-coupled receptor 65 (214467_at), score: 0.53 GPSM2G-protein signaling modulator 2 (AGS3-like, C. elegans) (221922_at), score: 0.59 GRSF1G-rich RNA sequence binding factor 1 (201501_s_at), score: -0.56 GTF2A1Lgeneral transcription factor IIA, 1-like (213413_at), score: 0.62 GTF2F2general transcription factor IIF, polypeptide 2, 30kDa (209595_at), score: -0.57 H1F0H1 histone family, member 0 (208886_at), score: 0.7 H1FXH1 histone family, member X (204805_s_at), score: 0.76 HAB1B1 for mucin (215778_x_at), score: -0.72 HAMPhepcidin antimicrobial peptide (220491_at), score: -0.63 HDGFhepatoma-derived growth factor (high-mobility group protein 1-like) (200896_x_at), score: 0.71 HGFhepatocyte growth factor (hepapoietin A; scatter factor) (209960_at), score: 0.74 HISPPD1histidine acid phosphatase domain containing 1 (203253_s_at), score: -0.55 HIST1H1Chistone cluster 1, H1c (209398_at), score: 0.63 HIST1H4Ghistone cluster 1, H4g (208551_at), score: -0.53 HLA-DQB1major histocompatibility complex, class II, DQ beta 1 (211654_x_at), score: -0.57 HMG20Bhigh-mobility group 20B (210719_s_at), score: 0.58 HMGA1high mobility group AT-hook 1 (210457_x_at), score: 0.76 HNRNPUL1heterogeneous nuclear ribonucleoprotein U-like 1 (209675_s_at), score: 0.65 HSD17B6hydroxysteroid (17-beta) dehydrogenase 6 homolog (mouse) (37512_at), score: -0.9 HSPA12Aheat shock 70kDa protein 12A (214434_at), score: 0.69 HSPA4Lheat shock 70kDa protein 4-like (205543_at), score: 0.61 HSPA9heat shock 70kDa protein 9 (mortalin) (200690_at), score: -0.54 IDUAiduronidase, alpha-L- (205059_s_at), score: 0.56 IFI30interferon, gamma-inducible protein 30 (201422_at), score: 0.56 IFRD1interferon-related developmental regulator 1 (202146_at), score: -0.53 IGHMBP2immunoglobulin mu binding protein 2 (215980_s_at), score: -0.55 INAinternexin neuronal intermediate filament protein, alpha (204465_s_at), score: 0.56 INE1inactivation escape 1 (non-protein coding) (207252_at), score: -0.6 INTS1integrator complex subunit 1 (212212_s_at), score: 0.53 IRF1interferon regulatory factor 1 (202531_at), score: 0.68 IRF2interferon regulatory factor 2 (203275_at), score: 0.55 ITGA1integrin, alpha 1 (214660_at), score: -0.7 ITGAVintegrin, alpha V (vitronectin receptor, alpha polypeptide, antigen CD51) (202351_at), score: -0.6 JAK2Janus kinase 2 (a protein tyrosine kinase) (205842_s_at), score: 0.53 JUNDjun D proto-oncogene (203751_x_at), score: 0.62 JUPjunction plakoglobin (201015_s_at), score: 0.83 KAL1Kallmann syndrome 1 sequence (205206_at), score: -1 KCNG1potassium voltage-gated channel, subfamily G, member 1 (214595_at), score: -0.54 KCTD15potassium channel tetramerisation domain containing 15 (218553_s_at), score: -0.64 KIAA0562KIAA0562 (204075_s_at), score: -0.64 KIAA1305KIAA1305 (220911_s_at), score: 0.76 KIF20Akinesin family member 20A (218755_at), score: 0.57 KLHL23kelch-like 23 (Drosophila) (213610_s_at), score: -0.72 KPNA6karyopherin alpha 6 (importin alpha 7) (212101_at), score: 0.55 LARP4La ribonucleoprotein domain family, member 4 (214155_s_at), score: -0.64 LARSleucyl-tRNA synthetase (217810_x_at), score: -0.64 LEPREL2leprecan-like 2 (204854_at), score: 0.55 LIG1ligase I, DNA, ATP-dependent (202726_at), score: 0.56 LIN7Blin-7 homolog B (C. elegans) (219760_at), score: -0.58 LLGL2lethal giant larvae homolog 2 (Drosophila) (203713_s_at), score: -0.63 LMNB2lamin B2 (216952_s_at), score: 0.57 LOC100129500hypothetical protein LOC100129500 (212884_x_at), score: -0.6 LOC100132540similar to LOC339047 protein (214870_x_at), score: 0.56 LOC100133105hypothetical protein LOC100133105 (214237_x_at), score: -0.61 LOC128192similar to peptidyl-Pro cis trans isomerase (217346_at), score: -0.53 LOC339047hypothetical protein LOC339047 (221501_x_at), score: 0.58 LOC391132similar to hCG2041276 (216177_at), score: -0.72 LOC440434hypothetical protein FLJ11822 (215090_x_at), score: 0.7 LPGAT1lysophosphatidylglycerol acyltransferase 1 (202651_at), score: -0.65 LRDDleucine-rich repeats and death domain containing (221640_s_at), score: 0.65 LRP2low density lipoprotein-related protein 2 (205710_at), score: 0.76 LRP8low density lipoprotein receptor-related protein 8, apolipoprotein e receptor (205282_at), score: -0.83 LRRN3leucine rich repeat neuronal 3 (209841_s_at), score: -0.53 MAFGv-maf musculoaponeurotic fibrosarcoma oncogene homolog G (avian) (204970_s_at), score: -0.54 MAN1C1mannosidase, alpha, class 1C, member 1 (218918_at), score: 0.58 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: 0.63 MAP2K2mitogen-activated protein kinase kinase 2 (213490_s_at), score: 0.53 MAP2K5mitogen-activated protein kinase kinase 5 (211370_s_at), score: 0.55 MAP3K6mitogen-activated protein kinase kinase kinase 6 (219278_at), score: 0.61 MAP7D1MAP7 domain containing 1 (217943_s_at), score: 0.53 MAPKAPK3mitogen-activated protein kinase-activated protein kinase 3 (202788_at), score: 0.65 MAST2microtubule associated serine/threonine kinase 2 (211593_s_at), score: 0.61 MAST4microtubule associated serine/threonine kinase family member 4 (40016_g_at), score: 0.62 MDM2Mdm2 p53 binding protein homolog (mouse) (205386_s_at), score: 0.79 MED15mediator complex subunit 15 (222175_s_at), score: 0.54 MFAP3Lmicrofibrillar-associated protein 3-like (205442_at), score: -0.68 MGAT1mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase (201126_s_at), score: 0.58 MINK1misshapen-like kinase 1 (zebrafish) (214246_x_at), score: 0.6 MLL4myeloid/lymphoid or mixed-lineage leukemia 4 (203419_at), score: 0.55 MMP10matrix metallopeptidase 10 (stromelysin 2) (205680_at), score: -0.71 MMP16matrix metallopeptidase 16 (membrane-inserted) (207012_at), score: -0.55 MOCS1molybdenum cofactor synthesis 1 (211673_s_at), score: -0.56 MPP1membrane protein, palmitoylated 1, 55kDa (202974_at), score: 0.73 MRASmuscle RAS oncogene homolog (206538_at), score: -0.54 MRPS6mitochondrial ribosomal protein S6 (213167_s_at), score: 0.61 MSTO1misato homolog 1 (Drosophila) (218296_x_at), score: -0.61 MTMR11myotubularin related protein 11 (205076_s_at), score: 0.55 MUC1mucin 1, cell surface associated (207847_s_at), score: 0.62 MUL1mitochondrial E3 ubiquitin ligase 1 (218246_at), score: 0.61 MYL4myosin, light chain 4, alkali; atrial, embryonic (210088_x_at), score: -0.7 NARG1LNMDA receptor regulated 1-like (219378_at), score: -0.63 NBL1neuroblastoma, suppression of tumorigenicity 1 (37005_at), score: 0.6 NCALDneurocalcin delta (211685_s_at), score: -0.64 NCAPD2non-SMC condensin I complex, subunit D2 (201774_s_at), score: 0.62 NCKAP1NCK-associated protein 1 (217465_at), score: -0.59 NDPNorrie disease (pseudoglioma) (206022_at), score: 0.84 NDUFA2NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2, 8kDa (213550_s_at), score: 0.55 NEFLneurofilament, light polypeptide (221805_at), score: -0.7 NEFMneurofilament, medium polypeptide (205113_at), score: -0.96 NEO1neogenin homolog 1 (chicken) (204321_at), score: 0.7 NF1neurofibromin 1 (211094_s_at), score: 0.78 NFATC4nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 (205897_at), score: 0.62 NFKBIAnuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha (201502_s_at), score: 0.76 NFYCnuclear transcription factor Y, gamma (202215_s_at), score: 0.56 NMIN-myc (and STAT) interactor (203964_at), score: 0.54 NOL10nucleolar protein 10 (218591_s_at), score: -0.64 NOVA2neuro-oncological ventral antigen 2 (206477_s_at), score: -0.59 NOX4NADPH oxidase 4 (219773_at), score: -0.64 NPIPnuclear pore complex interacting protein (204538_x_at), score: 0.65 NRXN3neurexin 3 (205795_at), score: -0.62 NUDT1nudix (nucleoside diphosphate linked moiety X)-type motif 1 (204766_s_at), score: 0.55 NUP160nucleoporin 160kDa (212709_at), score: -0.55 NUPL2nucleoporin like 2 (204003_s_at), score: -0.7 NXPH3neurexophilin 3 (221991_at), score: -0.67 OAS22'-5'-oligoadenylate synthetase 2, 69/71kDa (204972_at), score: 0.67 OAS32'-5'-oligoadenylate synthetase 3, 100kDa (218400_at), score: 0.75 OBFC2Boligonucleotide/oligosaccharide-binding fold containing 2B (218903_s_at), score: 0.55 OLFML2Bolfactomedin-like 2B (213125_at), score: 0.99 OLR1oxidized low density lipoprotein (lectin-like) receptor 1 (210004_at), score: -0.73 OPCMLopioid binding protein/cell adhesion molecule-like (214111_at), score: -0.73 OSTM1osteopetrosis associated transmembrane protein 1 (218196_at), score: -0.73 P2RY5purinergic receptor P2Y, G-protein coupled, 5 (218589_at), score: 0.66 PAK1p21 protein (Cdc42/Rac)-activated kinase 1 (209615_s_at), score: 0.69 PARNpoly(A)-specific ribonuclease (deadenylation nuclease) (203905_at), score: 0.58 PARVBparvin, beta (204629_at), score: 0.59 PBX3pre-B-cell leukemia homeobox 3 (204082_at), score: 0.56 PCDH9protocadherin 9 (219737_s_at), score: -0.94 PCDHG@protocadherin gamma cluster (215836_s_at), score: 0.53 PCDHGA1protocadherin gamma subfamily A, 1 (209079_x_at), score: 0.53 PCDHGA11protocadherin gamma subfamily A, 11 (211876_x_at), score: 0.58 PCDHGA3protocadherin gamma subfamily A, 3 (216352_x_at), score: 0.62 PCYT2phosphate cytidylyltransferase 2, ethanolamine (209577_at), score: 0.68 PDE5Aphosphodiesterase 5A, cGMP-specific (206757_at), score: 0.85 PDGFRAplatelet-derived growth factor receptor, alpha polypeptide (203131_at), score: 0.57 PHF7PHD finger protein 7 (215622_x_at), score: -0.61 PHLDA1pleckstrin homology-like domain, family A, member 1 (217997_at), score: 0.55 PIGHphosphatidylinositol glycan anchor biosynthesis, class H (209625_at), score: -0.57 PIGLphosphatidylinositol glycan anchor biosynthesis, class L (213889_at), score: 0.63 PIP5K1Cphosphatidylinositol-4-phosphate 5-kinase, type I, gamma (212518_at), score: 0.65 PITX1paired-like homeodomain 1 (208502_s_at), score: 0.7 PKN1protein kinase N1 (202161_at), score: 0.59 PLAUplasminogen activator, urokinase (211668_s_at), score: 0.69 PLCD1phospholipase C, delta 1 (205125_at), score: 0.61 PLK1polo-like kinase 1 (Drosophila) (202240_at), score: 0.68 PMLpromyelocytic leukemia (206503_x_at), score: 0.55 POLR2Apolymerase (RNA) II (DNA directed) polypeptide A, 220kDa (202725_at), score: 0.61 POM121POM121 membrane glycoprotein (rat) (212178_s_at), score: 0.56 POM121L2POM121 membrane glycoprotein-like 2 (rat) (216582_at), score: -0.53 POMZP3POM (POM121 homolog, rat) and ZP3 fusion (204148_s_at), score: 0.63 PPARGperoxisome proliferator-activated receptor gamma (208510_s_at), score: 0.68 PRMT3protein arginine methyltransferase 3 (213320_at), score: -0.8 PTPROprotein tyrosine phosphatase, receptor type, O (211600_at), score: 0.65 PTTG3pituitary tumor-transforming 3 (208511_at), score: 0.66 PXNpaxillin (211823_s_at), score: 0.72 RAB1ARAB1A, member RAS oncogene family (213440_at), score: -0.82 RAB22ARAB22A, member RAS oncogene family (218360_at), score: 0.55 RAB33BRAB33B, member RAS oncogene family (221014_s_at), score: -0.6 RABGGTBRab geranylgeranyltransferase, beta subunit (213704_at), score: -0.54 RAG1AP1recombination activating gene 1 activating protein 1 (219125_s_at), score: -0.56 RASGRP3RAS guanyl releasing protein 3 (calcium and DAG-regulated) (205801_s_at), score: 0.65 RASSF2Ras association (RalGDS/AF-6) domain family member 2 (203185_at), score: 0.54 RBM15BRNA binding motif protein 15B (202689_at), score: 0.6 REREarginine-glutamic acid dipeptide (RE) repeats (200940_s_at), score: 0.79 RFX5regulatory factor X, 5 (influences HLA class II expression) (202963_at), score: 0.69 RIN1Ras and Rab interactor 1 (205211_s_at), score: 0.54 RNF11ring finger protein 11 (208924_at), score: -0.58 RNF121ring finger protein 121 (219021_at), score: -0.55 RNF220ring finger protein 220 (219988_s_at), score: 0.77 RNF44ring finger protein 44 (203286_at), score: 0.56 RNFT1ring finger protein, transmembrane 1 (221194_s_at), score: -0.55 ROR2receptor tyrosine kinase-like orphan receptor 2 (205578_at), score: 0.54 RP3-377H14.5hypothetical LOC285830 (222279_at), score: -0.53 RP4-691N24.1ninein-like (207705_s_at), score: 0.53 RP5-1000E10.4suppressor of IKK epsilon (221705_s_at), score: -0.67 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: -0.8 RPL39Lribosomal protein L39-like (210115_at), score: 0.55 RPLP2P1ribosomal protein, large P2, pseudogene 1 (216490_x_at), score: -0.66 RPRD2regulation of nuclear pre-mRNA domain containing 2 (212553_at), score: 0.64 RPS17P5ribosomal protein S17 pseudogene 5 (216348_at), score: -0.73 RPS25ribosomal protein S25 (200091_s_at), score: -0.54 RPS6KA4ribosomal protein S6 kinase, 90kDa, polypeptide 4 (204632_at), score: 0.54 RRP15ribosomal RNA processing 15 homolog (S. cerevisiae) (219037_at), score: -0.67 SAA4serum amyloid A4, constitutive (207096_at), score: -0.61 SAP30LSAP30-like (219129_s_at), score: 0.72 SAV1salvador homolog 1 (Drosophila) (218276_s_at), score: 0.75 SBNO2strawberry notch homolog 2 (Drosophila) (204166_at), score: 0.66 SCAMP4secretory carrier membrane protein 4 (213244_at), score: 0.89 SCARA3scavenger receptor class A, member 3 (219416_at), score: 0.61 SDAD1SDA1 domain containing 1 (218607_s_at), score: 0.53 SDF2L1stromal cell-derived factor 2-like 1 (218681_s_at), score: -0.63 SDPRserum deprivation response (phosphatidylserine binding protein) (218711_s_at), score: 0.74 SEC61A2Sec61 alpha 2 subunit (S. cerevisiae) (219499_at), score: 0.57 SEMA3Fsema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F (35666_at), score: -0.6 SEPHS1selenophosphate synthetase 1 (208939_at), score: 0.64 SERPINB9serpin peptidase inhibitor, clade B (ovalbumin), member 9 (209723_at), score: 0.57 SF1splicing factor 1 (208313_s_at), score: 0.79 SH2B3SH2B adaptor protein 3 (203320_at), score: 0.55 SH3GLB2SH3-domain GRB2-like endophilin B2 (218813_s_at), score: 0.62 SIGIRRsingle immunoglobulin and toll-interleukin 1 receptor (TIR) domain (52940_at), score: 0.62 SIGLEC6sialic acid binding Ig-like lectin 6 (210796_x_at), score: -0.63 SIRT3sirtuin (silent mating type information regulation 2 homolog) 3 (S. cerevisiae) (221562_s_at), score: 0.53 SIRT6sirtuin (silent mating type information regulation 2 homolog) 6 (S. cerevisiae) (219613_s_at), score: 0.66 SLC25A13solute carrier family 25, member 13 (citrin) (203775_at), score: -0.74 SLC2A4RGSLC2A4 regulator (218494_s_at), score: 0.57 SLC33A1solute carrier family 33 (acetyl-CoA transporter), member 1 (203165_s_at), score: -0.69 SLC39A14solute carrier family 39 (zinc transporter), member 14 (212110_at), score: -0.55 SLC39A7solute carrier family 39 (zinc transporter), member 7 (202667_s_at), score: -0.59 SLC43A3solute carrier family 43, member 3 (213113_s_at), score: 0.68 SLC4A4solute carrier family 4, sodium bicarbonate cotransporter, member 4 (203908_at), score: -0.54 SLC4A7solute carrier family 4, sodium bicarbonate cotransporter, member 7 (209884_s_at), score: -0.76 SLC5A5solute carrier family 5 (sodium iodide symporter), member 5 (211123_at), score: -0.72 SLC6A11solute carrier family 6 (neurotransmitter transporter, GABA), member 11 (207048_at), score: -0.65 SLMO2slowmo homolog 2 (Drosophila) (217851_s_at), score: -0.57 SMAD3SMAD family member 3 (218284_at), score: 0.8 SMAP1small ArfGAP 1 (218137_s_at), score: -0.55 SMG6Smg-6 homolog, nonsense mediated mRNA decay factor (C. elegans) (214940_s_at), score: 0.61 SMN1survival of motor neuron 1, telomeric (203852_s_at), score: -0.54 SNAP25synaptosomal-associated protein, 25kDa (202508_s_at), score: 0.78 SNX27sorting nexin family member 27 (221006_s_at), score: 0.55 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: 0.67 SORBS1sorbin and SH3 domain containing 1 (218087_s_at), score: -0.62 SPA17sperm autoantigenic protein 17 (205406_s_at), score: 0.59 SPG7spastic paraplegia 7 (pure and complicated autosomal recessive) (202104_s_at), score: 0.64 ST5suppression of tumorigenicity 5 (202440_s_at), score: 0.57 STAT6signal transducer and activator of transcription 6, interleukin-4 induced (201331_s_at), score: 0.53 STIM1stromal interaction molecule 1 (202764_at), score: 0.75 SULT1B1sulfotransferase family, cytosolic, 1B, member 1 (207601_at), score: -0.66 SYNE1spectrin repeat containing, nuclear envelope 1 (209447_at), score: -0.72 SYNJ1synaptojanin 1 (212990_at), score: -0.58 TAF13TAF13 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 18kDa (205966_at), score: -0.6 TAPBPTAP binding protein (tapasin) (208829_at), score: 0.57 TBCDtubulin folding cofactor D (211052_s_at), score: 0.54 TDGthymine-DNA glycosylase (203743_s_at), score: -0.57 TENC1tensin like C1 domain containing phosphatase (tensin 2) (212494_at), score: 0.65 TERF2telomeric repeat binding factor 2 (203611_at), score: 0.54 TFDP2transcription factor Dp-2 (E2F dimerization partner 2) (203588_s_at), score: 0.53 TGDSTDP-glucose 4,6-dehydratase (208249_s_at), score: -0.67 TGFBR2transforming growth factor, beta receptor II (70/80kDa) (207334_s_at), score: 0.6 THSD7Athrombospondin, type I, domain containing 7A (214920_at), score: 0.76 TIAF1TGFB1-induced anti-apoptotic factor 1 (202039_at), score: 0.7 TICAM2toll-like receptor adaptor molecule 2 (214658_at), score: -0.75 TLE3transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) (206472_s_at), score: 0.6 TLE4transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) (204872_at), score: -0.67 TLR3toll-like receptor 3 (206271_at), score: 0.55 TMED7transmembrane emp24 protein transport domain containing 7 (209404_s_at), score: -0.56 TMEM104transmembrane protein 104 (220097_s_at), score: -0.65 TMEM2transmembrane protein 2 (218113_at), score: -0.6 TMEM39Btransmembrane protein 39B (218770_s_at), score: 0.58 TMSB15Bthymosin beta 15B (214051_at), score: 0.68 TNFRSF10Ctumor necrosis factor receptor superfamily, member 10c, decoy without an intracellular domain (206222_at), score: 0.59 TOMM20translocase of outer mitochondrial membrane 20 homolog (yeast) (200662_s_at), score: -0.63 TP53tumor protein p53 (201746_at), score: 0.76 TRAF5TNF receptor-associated factor 5 (204352_at), score: -0.61 TRIM21tripartite motif-containing 21 (204804_at), score: 0.68 TRMT2ATRM2 tRNA methyltransferase 2 homolog A (S. cerevisiae) (91617_at), score: -0.75 TRPC4transient receptor potential cation channel, subfamily C, member 4 (220817_at), score: -0.62 TSC2tuberous sclerosis 2 (215735_s_at), score: 0.6 TSKUtsukushin (218245_at), score: 0.74 TSPAN31tetraspanin 31 (203227_s_at), score: -0.54 TTC13tetratricopeptide repeat domain 13 (219481_at), score: -0.57 TUBA4Btubulin, alpha 4b (pseudogene) (207490_at), score: -0.67 TXLNAtaxilin alpha (212300_at), score: 0.57 UBAC1UBA domain containing 1 (202151_s_at), score: 0.55 UBTD1ubiquitin domain containing 1 (219172_at), score: 0.68 UGDHUDP-glucose dehydrogenase (203343_at), score: -0.59 ULBP2UL16 binding protein 2 (221291_at), score: -0.67 ULK1unc-51-like kinase 1 (C. elegans) (209333_at), score: 0.58 USF2upstream transcription factor 2, c-fos interacting (202152_x_at), score: 0.56 VEGFBvascular endothelial growth factor B (203683_s_at), score: 0.69 VPS13Cvacuolar protein sorting 13 homolog C (S. cerevisiae) (218396_at), score: -0.55 VPS37Bvacuolar protein sorting 37 homolog B (S. cerevisiae) (221704_s_at), score: 0.65 VWA5Avon Willebrand factor A domain containing 5A (210102_at), score: 0.59 WBP2WW domain binding protein 2 (209117_at), score: 0.54 WDR4WD repeat domain 4 (221632_s_at), score: -0.65 WDR42AWD repeat domain 42A (202249_s_at), score: 0.65 WDR6WD repeat domain 6 (217734_s_at), score: 0.83 WWTR1WW domain containing transcription regulator 1 (202132_at), score: -0.55 XAB2XPA binding protein 2 (218110_at), score: 0.54 YAF2YY1 associated factor 2 (206238_s_at), score: -0.58 YES1v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 (202932_at), score: 0.8 YPEL1yippee-like 1 (Drosophila) (206063_x_at), score: -0.67 YTHDC2YTH domain containing 2 (213077_at), score: -0.6 ZAKsterile alpha motif and leucine zipper containing kinase AZK (218833_at), score: -0.54 ZBTB20zinc finger and BTB domain containing 20 (205383_s_at), score: 0.57 ZCCHC10zinc finger, CCHC domain containing 10 (221193_s_at), score: -0.65 ZDHHC4zinc finger, DHHC-type containing 4 (220261_s_at), score: 0.54 ZDHHC7zinc finger, DHHC-type containing 7 (218606_at), score: 0.7 ZFYVE16zinc finger, FYVE domain containing 16 (203651_at), score: -0.53 ZFYVE26zinc finger, FYVE domain containing 26 (37943_at), score: -0.53 ZNF23zinc finger protein 23 (KOX 16) (213934_s_at), score: -0.57 ZNF281zinc finger protein 281 (218401_s_at), score: -0.59 ZNF395zinc finger protein 395 (218149_s_at), score: 0.56 ZNF580zinc finger protein 580 (220748_s_at), score: 0.55 ZNF706zinc finger protein 706 (218059_at), score: 0.64
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486171.cel | 27 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486311.cel | 34 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485791.cel | 8 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |