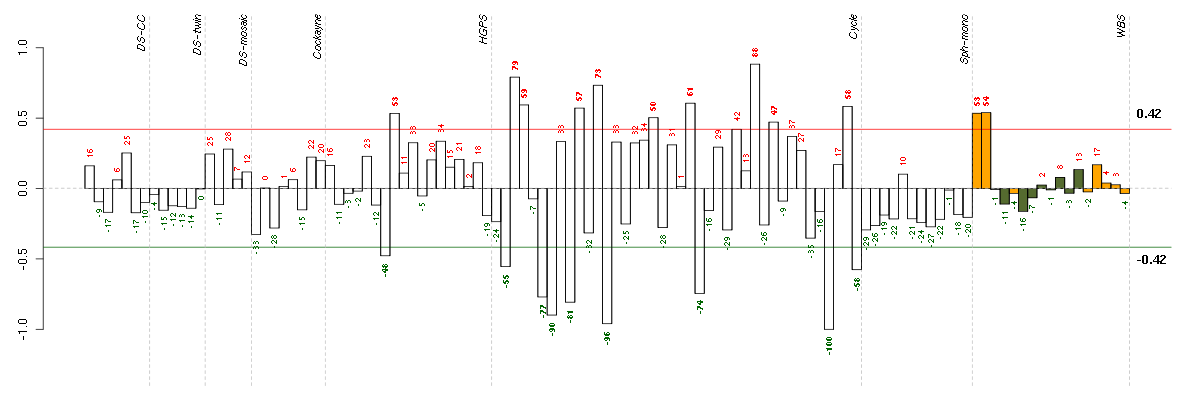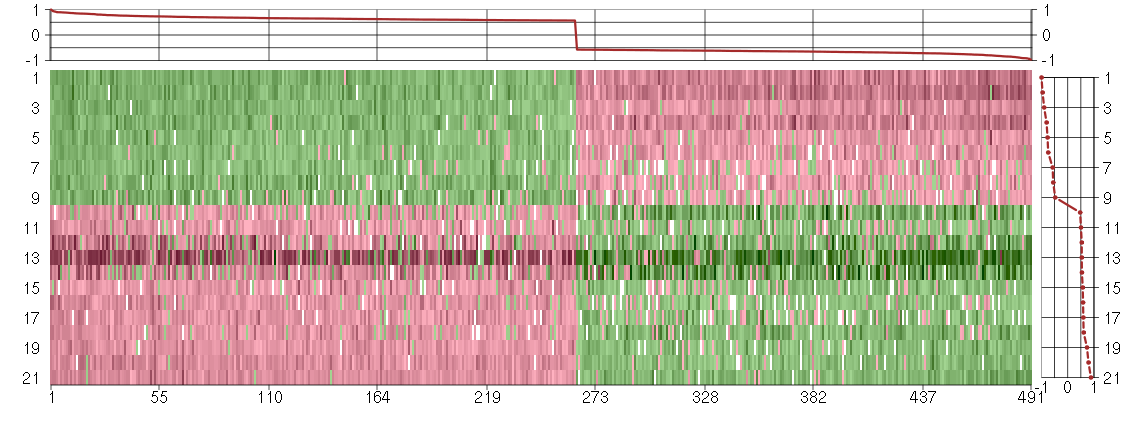

Under-expression is coded with green,
over-expression with red color.

signal transduction
The cascade of processes by which a signal interacts with a receptor, causing a change in the level or activity of a second messenger or other downstream target, and ultimately effecting a change in the functioning of the cell.
cell communication
Any process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
intracellular signaling cascade
A series of reactions within the cell that occur as a result of a single trigger reaction or compound.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
positive regulation of kinase activity
Any process that activates or increases the frequency, rate or extent of kinase activity, the catalysis of the transfer of a phosphate group, usually from ATP, to a substrate molecule.
positive regulation of catalytic activity
Any process that activates or increases the activity of an enzyme.
regulation of MAP kinase activity
Any process that modulates the frequency, rate or extent of MAP kinase activity.
positive regulation of MAP kinase activity
Any process that activates or increases the frequency, rate or extent of MAP kinase activity.
regulation of JUN kinase activity
Any process that modulates the frequency, rate or extent of JUN kinase activity.
positive regulation of JUN kinase activity
Any process that activates or increases the frequency, rate or extent of JUN kinase activity.
regulation of kinase activity
Any process that modulates the frequency, rate or extent of kinase activity, the catalysis of the transfer of a phosphate group, usually from ATP, to a substrate molecule.
regulation of protein kinase activity
Any process that modulates the frequency, rate or extent of protein kinase activity.
positive regulation of protein kinase activity
Any process that activates or increases the frequency, rate or extent of protein kinase activity.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of catalytic activity
Any process that modulates the activity of an enzyme.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of transferase activity
Any process that modulates the frequency, rate or extent of transferase activity, the catalysis of the transfer of a group, e.g. a methyl group, glycosyl group, acyl group, phosphorus-containing, or other groups, from one compound (generally regarded as the donor) to another compound (generally regarded as the acceptor). Transferase is the systematic name for any enzyme of EC class 2.
positive regulation of transferase activity
Any process that activates or increases the frequency, rate or extent of transferase activity, the catalysis of the transfer of a group, e.g. a methyl group, glycosyl group, acyl group, phosphorus-containing, or other groups, from a donor compound to an acceptor.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
regulation of molecular function
Any process that modulates the frequency, rate or extent of molecular functions. Molecular functions are elemental biological activities occurring at the molecular level, such as catalysis or binding.
all
This term is the most general term possible
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
signal transduction
The cascade of processes by which a signal interacts with a receptor, causing a change in the level or activity of a second messenger or other downstream target, and ultimately effecting a change in the functioning of the cell.
positive regulation of transferase activity
Any process that activates or increases the frequency, rate or extent of transferase activity, the catalysis of the transfer of a group, e.g. a methyl group, glycosyl group, acyl group, phosphorus-containing, or other groups, from a donor compound to an acceptor.
positive regulation of kinase activity
Any process that activates or increases the frequency, rate or extent of kinase activity, the catalysis of the transfer of a phosphate group, usually from ATP, to a substrate molecule.
positive regulation of protein kinase activity
Any process that activates or increases the frequency, rate or extent of protein kinase activity.
positive regulation of MAP kinase activity
Any process that activates or increases the frequency, rate or extent of MAP kinase activity.
positive regulation of JUN kinase activity
Any process that activates or increases the frequency, rate or extent of JUN kinase activity.


AAASachalasia, adrenocortical insufficiency, alacrimia (Allgrove, triple-A) (218075_at), score: 0.65 ABCD1ATP-binding cassette, sub-family D (ALD), member 1 (205142_x_at), score: 0.78 ABCE1ATP-binding cassette, sub-family E (OABP), member 1 (201873_s_at), score: -0.64 ABL1c-abl oncogene 1, receptor tyrosine kinase (202123_s_at), score: 0.58 ABLIM1actin binding LIM protein 1 (200965_s_at), score: 0.61 ACAP2ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 (212476_at), score: -0.68 ACN9ACN9 homolog (S. cerevisiae) (218981_at), score: -0.63 ACSL4acyl-CoA synthetase long-chain family member 4 (202422_s_at), score: -0.81 ACTR3BARP3 actin-related protein 3 homolog B (yeast) (218868_at), score: -0.69 ADCK2aarF domain containing kinase 2 (221893_s_at), score: 0.89 AGAP1ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 (204066_s_at), score: 0.62 AGPAT11-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) (215535_s_at), score: 0.82 AHI1Abelson helper integration site 1 (221569_at), score: -0.64 AKAP2A kinase (PRKA) anchor protein 2 (202759_s_at), score: 0.68 ALDH1L1aldehyde dehydrogenase 1 family, member L1 (215798_at), score: -0.7 ANXA2P1annexin A2 pseudogene 1 (210876_at), score: -0.59 AP2A2adaptor-related protein complex 2, alpha 2 subunit (212159_x_at), score: 0.69 ARFGAP3ADP-ribosylation factor GTPase activating protein 3 (202211_at), score: -0.64 ARHGDIARho GDP dissociation inhibitor (GDI) alpha (213606_s_at), score: 0.66 ARSAarylsulfatase A (204443_at), score: 0.79 ATF2activating transcription factor 2 (212984_at), score: -0.74 ATF5activating transcription factor 5 (204999_s_at), score: 0.71 ATG9AATG9 autophagy related 9 homolog A (S. cerevisiae) (202492_at), score: 0.58 ATN1atrophin 1 (40489_at), score: 0.75 ATP2C1ATPase, Ca++ transporting, type 2C, member 1 (211137_s_at), score: -0.68 ATP8B1ATPase, class I, type 8B, member 1 (214594_x_at), score: -0.66 AXLAXL receptor tyrosine kinase (202685_s_at), score: 0.57 AZI25-azacytidine induced 2 (218043_s_at), score: -0.6 B3GAT3beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) (203452_at), score: 0.59 BAP1BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) (201419_at), score: 0.67 BAT2HLA-B associated transcript 2 (212081_x_at), score: 0.88 BAT2LHLA-B associated transcript 2-like (212068_s_at), score: 0.66 BAT3HLA-B associated transcript 3 (213318_s_at), score: 0.7 BCKDKbranched chain ketoacid dehydrogenase kinase (202030_at), score: 0.57 BCL2L1BCL2-like 1 (215037_s_at), score: 0.61 BTBD2BTB (POZ) domain containing 2 (207722_s_at), score: 0.92 C12orf29chromosome 12 open reading frame 29 (213701_at), score: -0.6 C14orf104chromosome 14 open reading frame 104 (219166_at), score: -0.59 C14orf138chromosome 14 open reading frame 138 (218940_at), score: -0.61 C14orf147chromosome 14 open reading frame 147 (212460_at), score: -0.61 C14orf94chromosome 14 open reading frame 94 (218383_at), score: 0.72 C15orf33chromosome 15 open reading frame 33 (216411_s_at), score: -0.63 C16orf45chromosome 16 open reading frame 45 (212736_at), score: 0.65 C16orf57chromosome 16 open reading frame 57 (218060_s_at), score: 0.61 C19orf61chromosome 19 open reading frame 61 (221335_x_at), score: 0.68 C3orf64chromosome 3 open reading frame 64 (221935_s_at), score: -0.58 C4orf43chromosome 4 open reading frame 43 (218513_at), score: -0.88 C5orf28chromosome 5 open reading frame 28 (219029_at), score: -0.61 C5orf44chromosome 5 open reading frame 44 (218674_at), score: -0.9 C6orf211chromosome 6 open reading frame 211 (218195_at), score: -0.64 C7orf64chromosome 7 open reading frame 64 (221596_s_at), score: -0.62 C9orf82chromosome 9 open reading frame 82 (219276_x_at), score: -0.66 CABIN1calcineurin binding protein 1 (37652_at), score: 0.74 CALB2calbindin 2 (205428_s_at), score: -0.68 CALCOCO1calcium binding and coiled-coil domain 1 (209002_s_at), score: 0.75 CAPN1calpain 1, (mu/I) large subunit (200752_s_at), score: 0.58 CAPN5calpain 5 (205166_at), score: 0.68 CASKIN2CASK interacting protein 2 (61297_at), score: 0.57 CASP7caspase 7, apoptosis-related cysteine peptidase (207181_s_at), score: -0.69 CBX4chromobox homolog 4 (Pc class homolog, Drosophila) (206724_at), score: 0.57 CCDC41coiled-coil domain containing 41 (219644_at), score: -0.63 CCDC76coiled-coil domain containing 76 (219130_at), score: -0.66 CD248CD248 molecule, endosialin (219025_at), score: 0.73 CDC25Bcell division cycle 25 homolog B (S. pombe) (201853_s_at), score: 0.63 CDC2L6cell division cycle 2-like 6 (CDK8-like) (212899_at), score: 0.58 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (217849_s_at), score: 0.89 CDC42EP1CDC42 effector protein (Rho GTPase binding) 1 (204693_at), score: 0.71 CDC42EP4CDC42 effector protein (Rho GTPase binding) 4 (214721_x_at), score: 0.62 CDC73cell division cycle 73, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae) (218578_at), score: -0.61 CDH13cadherin 13, H-cadherin (heart) (204726_at), score: -0.62 CENPTcentromere protein T (218148_at), score: 0.6 CHMP5chromatin modifying protein 5 (218085_at), score: -0.75 CHORDC1cysteine and histidine-rich domain (CHORD)-containing 1 (218566_s_at), score: -0.71 CHUKconserved helix-loop-helix ubiquitous kinase (209666_s_at), score: -0.76 CICcapicua homolog (Drosophila) (212784_at), score: 0.86 CITcitron (rho-interacting, serine/threonine kinase 21) (212801_at), score: 0.67 CIZ1CDKN1A interacting zinc finger protein 1 (205516_x_at), score: 0.88 CLIP2CAP-GLY domain containing linker protein 2 (211031_s_at), score: 0.6 CLIP3CAP-GLY domain containing linker protein 3 (212358_at), score: 0.57 CMPK1cytidine monophosphate (UMP-CMP) kinase 1, cytosolic (217870_s_at), score: -0.65 CNIH3cornichon homolog 3 (Drosophila) (214841_at), score: 0.6 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: 0.89 CRCPCGRP receptor component (203898_at), score: -0.64 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: 0.86 CScitrate synthase (208660_at), score: 0.63 CUL7cullin 7 (36084_at), score: 0.61 CYP3A4cytochrome P450, family 3, subfamily A, polypeptide 4 (205998_x_at), score: -0.72 CYP7B1cytochrome P450, family 7, subfamily B, polypeptide 1 (207386_at), score: 0.6 DAXXdeath-domain associated protein (201763_s_at), score: 0.63 DBN1drebrin 1 (217025_s_at), score: 0.64 DCP2DCP2 decapping enzyme homolog (S. cerevisiae) (212919_at), score: -0.69 DDAH2dimethylarginine dimethylaminohydrolase 2 (215537_x_at), score: 0.58 DDB2damage-specific DNA binding protein 2, 48kDa (203409_at), score: 0.58 DENND4CDENN/MADD domain containing 4C (205684_s_at), score: -0.65 DENND5BDENN/MADD domain containing 5B (215058_at), score: -0.7 DERL2Der1-like domain family, member 2 (218333_at), score: -0.63 DGCR2DiGeorge syndrome critical region gene 2 (214198_s_at), score: -0.59 DKFZP564O0823DKFZP564O0823 protein (204687_at), score: -0.61 DMPKdystrophia myotonica-protein kinase (37996_s_at), score: 0.68 DNAJB9DnaJ (Hsp40) homolog, subfamily B, member 9 (202843_at), score: -0.85 DNAJC24DnaJ (Hsp40) homolog, subfamily C, member 24 (213853_at), score: -0.67 DNAJC6DnaJ (Hsp40) homolog, subfamily C, member 6 (204720_s_at), score: -0.62 DNM2dynamin 2 (202253_s_at), score: 0.69 DNPEPaspartyl aminopeptidase (201937_s_at), score: 0.64 DOCK9dedicator of cytokinesis 9 (212538_at), score: -0.59 DOPEY1dopey family member 1 (40612_at), score: -0.84 DRG2developmentally regulated GTP binding protein 2 (203267_s_at), score: 0.58 DSPPdentin sialophosphoprotein (221681_s_at), score: -0.7 DULLARDdullard homolog (Xenopus laevis) (200035_at), score: 0.58 DUS2Ldihydrouridine synthase 2-like, SMM1 homolog (S. cerevisiae) (47105_at), score: -0.57 EAF2ELL associated factor 2 (219551_at), score: -0.65 ECM1extracellular matrix protein 1 (209365_s_at), score: 0.71 EDEM1ER degradation enhancer, mannosidase alpha-like 1 (203279_at), score: -0.59 EDNRAendothelin receptor type A (204464_s_at), score: -0.65 EEA1early endosome antigen 1 (204840_s_at), score: -0.61 EFHD2EF-hand domain family, member D2 (217992_s_at), score: 0.59 EHBP1L1EH domain binding protein 1-like 1 (221755_at), score: 0.76 EHD2EH-domain containing 2 (221870_at), score: 0.58 EHMT2euchromatic histone-lysine N-methyltransferase 2 (202326_at), score: 0.6 EIF2AK3eukaryotic translation initiation factor 2-alpha kinase 3 (218696_at), score: -0.59 EIF3Meukaryotic translation initiation factor 3, subunit M (202231_at), score: -0.61 EIF4Eeukaryotic translation initiation factor 4E (201436_at), score: -0.81 EIF4G3eukaryotic translation initiation factor 4 gamma, 3 (201936_s_at), score: -0.67 EIF5Beukaryotic translation initiation factor 5B (214314_s_at), score: -0.61 ENGASEendo-beta-N-acetylglucosaminidase (220349_s_at), score: -0.6 ENO2enolase 2 (gamma, neuronal) (201313_at), score: 0.68 ENTPD1ectonucleoside triphosphate diphosphohydrolase 1 (209473_at), score: -0.67 EPHA4EPH receptor A4 (206114_at), score: -0.75 EPS15epidermal growth factor receptor pathway substrate 15 (217887_s_at), score: -0.66 ERBB2IPerbb2 interacting protein (217941_s_at), score: -0.71 EVI5ecotropic viral integration site 5 (209717_at), score: -0.59 EVLEnah/Vasp-like (217838_s_at), score: 0.66 F8A1coagulation factor VIII-associated (intronic transcript) 1 (203274_at), score: 0.66 FAM115Afamily with sequence similarity 115, member A (212981_s_at), score: -0.64 FAM176Bfamily with sequence similarity 176, member B (220134_x_at), score: 0.61 FAM18Bfamily with sequence similarity 18, member B (218446_s_at), score: -0.91 FAM3Afamily with sequence similarity 3, member A (38043_at), score: 0.57 FAM3Cfamily with sequence similarity 3, member C (201889_at), score: -0.59 FAM62Afamily with sequence similarity 62 (C2 domain containing), member A (208858_s_at), score: 0.59 FAM65Afamily with sequence similarity 65, member A (45749_at), score: 0.62 FBXL14F-box and leucine-rich repeat protein 14 (213145_at), score: 0.68 FBXO17F-box protein 17 (220233_at), score: 0.57 FBXO2F-box protein 2 (219305_x_at), score: -0.73 FCER1GFc fragment of IgE, high affinity I, receptor for; gamma polypeptide (204232_at), score: -0.69 FCF1FCF1 small subunit (SSU) processome component homolog (S. cerevisiae) (219927_at), score: -0.6 FHOD1formin homology 2 domain containing 1 (218530_at), score: 0.61 FKBP8FK506 binding protein 8, 38kDa (208255_s_at), score: 0.65 FKRPfukutin related protein (219853_at), score: 0.59 FKSG2apoptosis inhibitor (208588_at), score: -0.59 FLJ12529pre-mRNA cleavage factor I, 59 kDa subunit (217866_at), score: 0.62 FLOT1flotillin 1 (210142_x_at), score: 0.69 FMR1fragile X mental retardation 1 (215245_x_at), score: -0.58 FNDC4fibronectin type III domain containing 4 (218843_at), score: 0.58 FOSL1FOS-like antigen 1 (204420_at), score: 0.66 FOXC2forkhead box C2 (MFH-1, mesenchyme forkhead 1) (214520_at), score: 0.68 FOXK2forkhead box K2 (203064_s_at), score: 0.7 FREQfrequenin homolog (Drosophila) (218266_s_at), score: 0.58 FRG1FSHD region gene 1 (204145_at), score: -0.6 FUBP3far upstream element (FUSE) binding protein 3 (212824_at), score: -0.7 FUT2fucosyltransferase 2 (secretor status included) (210608_s_at), score: -0.59 G3BP2GTPase activating protein (SH3 domain) binding protein 2 (208841_s_at), score: -0.73 GABARAPL3GABA(A) receptors associated protein like 3 (pseudogene) (211458_s_at), score: -0.62 GBLG protein beta subunit-like (220587_s_at), score: 0.58 GFOD1glucose-fructose oxidoreductase domain containing 1 (219821_s_at), score: -0.8 GGA3golgi associated, gamma adaptin ear containing, ARF binding protein 3 (209411_s_at), score: 0.65 GHRgrowth hormone receptor (205498_at), score: -0.65 GK3Pglycerol kinase 3 pseudogene (215966_x_at), score: -0.64 GMCL1germ cell-less homolog 1 (Drosophila) (218458_at), score: -0.71 GNAI2guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2 (201040_at), score: 0.57 GNPTABN-acetylglucosamine-1-phosphate transferase, alpha and beta subunits (212959_s_at), score: -0.74 GOLT1Bgolgi transport 1 homolog B (S. cerevisiae) (218193_s_at), score: -0.67 GRINAglutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) (212090_at), score: 0.72 GTF2F2general transcription factor IIF, polypeptide 2, 30kDa (209595_at), score: -0.7 H1F0H1 histone family, member 0 (208886_at), score: 0.63 H1FXH1 histone family, member X (204805_s_at), score: 0.77 HAB1B1 for mucin (215778_x_at), score: -0.61 HBS1LHBS1-like (S. cerevisiae) (209314_s_at), score: -0.68 HDAC5histone deacetylase 5 (202455_at), score: 0.63 HDAC6histone deacetylase 6 (206846_s_at), score: 0.57 HDGFhepatoma-derived growth factor (high-mobility group protein 1-like) (200896_x_at), score: 0.68 HGShepatocyte growth factor-regulated tyrosine kinase substrate (210428_s_at), score: 0.57 HISPPD1histidine acid phosphatase domain containing 1 (203253_s_at), score: -0.78 HMG20Bhigh-mobility group 20B (210719_s_at), score: 0.63 HMGA1high mobility group AT-hook 1 (210457_x_at), score: 0.84 HNRNPH2heterogeneous nuclear ribonucleoprotein H2 (H') (201132_at), score: -0.85 HNRNPUL1heterogeneous nuclear ribonucleoprotein U-like 1 (209675_s_at), score: 0.74 HPNhepsin (204934_s_at), score: -0.65 HPS6Hermansky-Pudlak syndrome 6 (219052_at), score: 0.6 HSD17B6hydroxysteroid (17-beta) dehydrogenase 6 homolog (mouse) (37512_at), score: -0.91 HSF1heat shock transcription factor 1 (202344_at), score: 0.65 HSPA9heat shock 70kDa protein 9 (mortalin) (200690_at), score: -0.58 HUS1HUS1 checkpoint homolog (S. pombe) (217618_x_at), score: -0.63 IBTKinhibitor of Bruton agammaglobulinemia tyrosine kinase (210970_s_at), score: -0.67 IDUAiduronidase, alpha-L- (205059_s_at), score: 0.78 IFRD1interferon-related developmental regulator 1 (202146_at), score: -0.65 IGHMBP2immunoglobulin mu binding protein 2 (215980_s_at), score: -0.68 IL22RA1interleukin 22 receptor, alpha 1 (220056_at), score: -0.63 IMPACTImpact homolog (mouse) (218637_at), score: -0.64 INE1inactivation escape 1 (non-protein coding) (207252_at), score: -0.58 INPP1inositol polyphosphate-1-phosphatase (202794_at), score: 0.65 INTS1integrator complex subunit 1 (212212_s_at), score: 0.59 ITGA1integrin, alpha 1 (214660_at), score: -0.68 ITGAVintegrin, alpha V (vitronectin receptor, alpha polypeptide, antigen CD51) (202351_at), score: -0.66 JMJD1Cjumonji domain containing 1C (221763_at), score: -0.59 JUNDjun D proto-oncogene (203751_x_at), score: 0.7 JUPjunction plakoglobin (201015_s_at), score: 0.7 KAL1Kallmann syndrome 1 sequence (205206_at), score: -0.91 KIAA0562KIAA0562 (204075_s_at), score: -0.75 KIAA1305KIAA1305 (220911_s_at), score: 0.65 KLHL23kelch-like 23 (Drosophila) (213610_s_at), score: -0.68 LARP4La ribonucleoprotein domain family, member 4 (214155_s_at), score: -0.86 LARSleucyl-tRNA synthetase (217810_x_at), score: -0.77 LEPREL2leprecan-like 2 (204854_at), score: 0.63 LIMS2LIM and senescent cell antigen-like domains 2 (220765_s_at), score: 0.58 LLGL2lethal giant larvae homolog 2 (Drosophila) (203713_s_at), score: -0.68 LMF2lipase maturation factor 2 (212682_s_at), score: 0.73 LOC100132540similar to LOC339047 protein (214870_x_at), score: 0.61 LOC128192similar to peptidyl-Pro cis trans isomerase (217346_at), score: -0.72 LOC339047hypothetical protein LOC339047 (221501_x_at), score: 0.62 LOC391132similar to hCG2041276 (216177_at), score: -0.82 LOC440434hypothetical protein FLJ11822 (215090_x_at), score: 0.72 LOC90379hypothetical protein BC002926 (221849_s_at), score: 0.65 LPGAT1lysophosphatidylglycerol acyltransferase 1 (202651_at), score: -0.84 LRDDleucine-rich repeats and death domain containing (221640_s_at), score: 0.63 LRP8low density lipoprotein receptor-related protein 8, apolipoprotein e receptor (205282_at), score: -0.76 LRRC40leucine rich repeat containing 40 (218577_at), score: -0.63 LRRN3leucine rich repeat neuronal 3 (209841_s_at), score: -0.67 LTBP3latent transforming growth factor beta binding protein 3 (219922_s_at), score: 0.6 MAGOH2mago-nashi homolog 2, proliferation-associated (Drosophila) (217693_x_at), score: -0.71 MAN1B1mannosidase, alpha, class 1B, member 1 (65884_at), score: 0.58 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: 0.93 MAP2microtubule-associated protein 2 (210015_s_at), score: -0.63 MAP2K2mitogen-activated protein kinase kinase 2 (213490_s_at), score: 0.65 MAP3K11mitogen-activated protein kinase kinase kinase 11 (203652_at), score: 0.59 MAP3K6mitogen-activated protein kinase kinase kinase 6 (219278_at), score: 0.84 MAP4microtubule-associated protein 4 (200836_s_at), score: 0.65 MAP7D1MAP7 domain containing 1 (217943_s_at), score: 0.79 MAPK1IP1Lmitogen-activated protein kinase 1 interacting protein 1-like (212499_s_at), score: -0.58 MAPK3mitogen-activated protein kinase 3 (212046_x_at), score: 0.66 MAPK7mitogen-activated protein kinase 7 (35617_at), score: 0.75 MAPKAPK3mitogen-activated protein kinase-activated protein kinase 3 (202788_at), score: 0.72 MAST2microtubule associated serine/threonine kinase 2 (211593_s_at), score: 0.69 MAST4microtubule associated serine/threonine kinase family member 4 (40016_g_at), score: 0.62 MATR3matrin 3 (200626_s_at), score: -0.61 MBOAT7membrane bound O-acyltransferase domain containing 7 (209179_s_at), score: 0.64 MCM9minichromosome maintenance complex component 9 (219673_at), score: -0.6 MED12mediator complex subunit 12 (216071_x_at), score: 0.6 MED15mediator complex subunit 15 (222175_s_at), score: 0.76 MED23mediator complex subunit 23 (218846_at), score: -0.62 MEN1multiple endocrine neoplasia I (202645_s_at), score: 0.62 METRNmeteorin, glial cell differentiation regulator (219051_x_at), score: 0.57 MEX3Cmex-3 homolog C (C. elegans) (218247_s_at), score: -0.65 MFAP3Lmicrofibrillar-associated protein 3-like (205442_at), score: -0.64 MGAT1mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase (201126_s_at), score: 0.7 MGAT4Bmannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme B (220189_s_at), score: 0.64 MINK1misshapen-like kinase 1 (zebrafish) (214246_x_at), score: 0.8 MLL4myeloid/lymphoid or mixed-lineage leukemia 4 (203419_at), score: 0.58 MMP10matrix metallopeptidase 10 (stromelysin 2) (205680_at), score: -0.68 MOSPD1motile sperm domain containing 1 (218853_s_at), score: -0.63 MPHOSPH10M-phase phosphoprotein 10 (U3 small nucleolar ribonucleoprotein) (212885_at), score: -0.58 MPP1membrane protein, palmitoylated 1, 55kDa (202974_at), score: 0.59 MTMR11myotubularin related protein 11 (205076_s_at), score: 0.6 MUC1mucin 1, cell surface associated (207847_s_at), score: 0.82 MUL1mitochondrial E3 ubiquitin ligase 1 (218246_at), score: 0.64 MYL4myosin, light chain 4, alkali; atrial, embryonic (210088_x_at), score: -0.71 MYO1Bmyosin IB (212364_at), score: -0.61 MYO9Bmyosin IXB (217297_s_at), score: 0.57 NARG1LNMDA receptor regulated 1-like (219378_at), score: -0.74 NBL1neuroblastoma, suppression of tumorigenicity 1 (37005_at), score: 0.67 NCKAP1NCK-associated protein 1 (217465_at), score: -0.8 NCOR2nuclear receptor co-repressor 2 (207760_s_at), score: 0.68 NDUFA5NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5, 13kDa (201304_at), score: -0.73 NEFLneurofilament, light polypeptide (221805_at), score: -0.58 NEFMneurofilament, medium polypeptide (205113_at), score: -0.78 NENFneuron derived neurotrophic factor (218407_x_at), score: 0.63 NF1neurofibromin 1 (211094_s_at), score: 0.73 NFATC4nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 (205897_at), score: 0.82 NFKBIAnuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha (201502_s_at), score: 0.76 NHLRC2NHL repeat containing 2 (219353_at), score: -0.63 NLGN4Yneuroligin 4, Y-linked (207703_at), score: -0.61 NOL10nucleolar protein 10 (218591_s_at), score: -0.63 NOVA2neuro-oncological ventral antigen 2 (206477_s_at), score: -0.66 NOX4NADPH oxidase 4 (219773_at), score: -0.59 NPDC1neural proliferation, differentiation and control, 1 (218086_at), score: 0.62 NPIPnuclear pore complex interacting protein (204538_x_at), score: 0.73 NPR2natriuretic peptide receptor B/guanylate cyclase B (atrionatriuretic peptide receptor B) (204310_s_at), score: 0.65 NUP160nucleoporin 160kDa (212709_at), score: -0.71 NUPL2nucleoporin like 2 (204003_s_at), score: -0.67 OAS32'-5'-oligoadenylate synthetase 3, 100kDa (218400_at), score: 0.59 OBFC2Boligonucleotide/oligosaccharide-binding fold containing 2B (218903_s_at), score: 0.6 OGDHoxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) (201282_at), score: 0.71 OLFML2Bolfactomedin-like 2B (213125_at), score: 0.71 OLR1oxidized low density lipoprotein (lectin-like) receptor 1 (210004_at), score: -0.77 OPCMLopioid binding protein/cell adhesion molecule-like (214111_at), score: -0.65 OSTM1osteopetrosis associated transmembrane protein 1 (218196_at), score: -0.76 PAK1p21 protein (Cdc42/Rac)-activated kinase 1 (209615_s_at), score: 0.73 PARVBparvin, beta (204629_at), score: 0.83 PBX3pre-B-cell leukemia homeobox 3 (204082_at), score: 0.62 PCDH9protocadherin 9 (219737_s_at), score: -0.8 PCDHG@protocadherin gamma cluster (215836_s_at), score: 0.73 PCDHGA1protocadherin gamma subfamily A, 1 (209079_x_at), score: 0.78 PCDHGA11protocadherin gamma subfamily A, 11 (211876_x_at), score: 0.68 PCDHGA3protocadherin gamma subfamily A, 3 (216352_x_at), score: 0.69 PCDHGC3protocadherin gamma subfamily C, 3 (211066_x_at), score: 0.66 PCTK1PCTAIRE protein kinase 1 (208824_x_at), score: 0.58 PDCLphosducin-like (204449_at), score: -0.61 PEX6peroxisomal biogenesis factor 6 (204545_at), score: 0.66 PHF7PHD finger protein 7 (215622_x_at), score: -0.63 PIGOphosphatidylinositol glycan anchor biosynthesis, class O (209998_at), score: 0.65 PINK1PTEN induced putative kinase 1 (209018_s_at), score: 0.68 PIP5K1Cphosphatidylinositol-4-phosphate 5-kinase, type I, gamma (212518_at), score: 0.85 PKN1protein kinase N1 (202161_at), score: 0.77 PLAUplasminogen activator, urokinase (211668_s_at), score: 0.64 PLCD1phospholipase C, delta 1 (205125_at), score: 0.8 PLD3phospholipase D family, member 3 (201050_at), score: 0.66 PLEC1plectin 1, intermediate filament binding protein 500kDa (216971_s_at), score: 0.57 PLSCR3phospholipid scramblase 3 (218828_at), score: 0.57 PNPLA2patatin-like phospholipase domain containing 2 (212705_x_at), score: 0.83 PNPLA6patatin-like phospholipase domain containing 6 (203718_at), score: 0.7 POLR2Apolymerase (RNA) II (DNA directed) polypeptide A, 220kDa (202725_at), score: 0.74 POM121POM121 membrane glycoprotein (rat) (212178_s_at), score: 0.74 POM121L2POM121 membrane glycoprotein-like 2 (rat) (216582_at), score: -0.62 POMZP3POM (POM121 homolog, rat) and ZP3 fusion (204148_s_at), score: 0.7 PPP1CBprotein phosphatase 1, catalytic subunit, beta isoform (201407_s_at), score: -0.69 PPP2R5Bprotein phosphatase 2, regulatory subunit B', beta isoform (635_s_at), score: 0.66 PRKCDprotein kinase C, delta (202545_at), score: 0.73 PRKD2protein kinase D2 (209282_at), score: 0.61 PRMT3protein arginine methyltransferase 3 (213320_at), score: -0.88 PTK2PTK2 protein tyrosine kinase 2 (208820_at), score: -0.57 PTOV1prostate tumor overexpressed 1 (212032_s_at), score: 0.69 PTPROprotein tyrosine phosphatase, receptor type, O (211600_at), score: 0.63 PVRL3poliovirus receptor-related 3 (213325_at), score: -0.63 PVT1Pvt1 oncogene (non-protein coding) (216249_at), score: -0.6 PXNpaxillin (211823_s_at), score: 0.84 RAB11BRAB11B, member RAS oncogene family (34478_at), score: 0.64 RAB1ARAB1A, member RAS oncogene family (213440_at), score: -0.86 RAB33BRAB33B, member RAS oncogene family (221014_s_at), score: -0.66 RAB7ARAB7A, member RAS oncogene family (211960_s_at), score: -0.63 RABGGTBRab geranylgeranyltransferase, beta subunit (213704_at), score: -0.69 RAD54L2RAD54-like 2 (S. cerevisiae) (213205_s_at), score: 0.6 RANBP3RAN binding protein 3 (210120_s_at), score: 0.59 RANBP6RAN binding protein 6 (213019_at), score: -0.57 RARGretinoic acid receptor, gamma (204189_at), score: 0.59 RBM15BRNA binding motif protein 15B (202689_at), score: 0.75 RBM42RNA binding motif protein 42 (205740_s_at), score: 0.6 RELAv-rel reticuloendotheliosis viral oncogene homolog A (avian) (209878_s_at), score: 0.66 REREarginine-glutamic acid dipeptide (RE) repeats (200940_s_at), score: 0.78 RHOGras homolog gene family, member G (rho G) (203175_at), score: 0.62 RIN1Ras and Rab interactor 1 (205211_s_at), score: 0.6 RNF11ring finger protein 11 (208924_at), score: -0.72 RNF220ring finger protein 220 (219988_s_at), score: 0.88 RNF40ring finger protein 40 (206845_s_at), score: 0.59 RP2retinitis pigmentosa 2 (X-linked recessive) (205191_at), score: -0.58 RP3-377H14.5hypothetical LOC285830 (222279_at), score: -0.64 RP5-1000E10.4suppressor of IKK epsilon (221705_s_at), score: -0.71 RP5-886K2.1neuronal thread protein AD7c-NTP (207953_at), score: -0.58 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: -0.83 RPL21P68ribosomal protein L21 pseudogene 68 (217340_at), score: -0.64 RPLP2P1ribosomal protein, large P2, pseudogene 1 (216490_x_at), score: -0.63 RPS10P11ribosomal protein S10 pseudogene 11 (216505_x_at), score: -0.58 RPS17P5ribosomal protein S17 pseudogene 5 (216348_at), score: -0.93 RPS25ribosomal protein S25 (200091_s_at), score: -0.74 RPS27Aribosomal protein S27a (217144_at), score: -0.64 RPS6KA4ribosomal protein S6 kinase, 90kDa, polypeptide 4 (204632_at), score: 0.65 RRP15ribosomal RNA processing 15 homolog (S. cerevisiae) (219037_at), score: -0.75 SAA4serum amyloid A4, constitutive (207096_at), score: -0.58 SACM1LSAC1 suppressor of actin mutations 1-like (yeast) (202797_at), score: -0.65 SAP30LSAP30-like (219129_s_at), score: 0.6 SAV1salvador homolog 1 (Drosophila) (218276_s_at), score: 0.58 SBF1SET binding factor 1 (39835_at), score: 0.62 SBNO2strawberry notch homolog 2 (Drosophila) (204166_at), score: 0.8 SCAMP4secretory carrier membrane protein 4 (213244_at), score: 0.87 SCAPSREBF chaperone (212329_at), score: 0.61 SEC62SEC62 homolog (S. cerevisiae) (208942_s_at), score: -0.62 SEMA3Fsema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F (35666_at), score: -0.58 SF1splicing factor 1 (208313_s_at), score: 0.66 SF3A2splicing factor 3a, subunit 2, 66kDa (37462_i_at), score: 0.65 SH3BP5SH3-domain binding protein 5 (BTK-associated) (201810_s_at), score: 0.59 SH3GLB2SH3-domain GRB2-like endophilin B2 (218813_s_at), score: 0.69 SHBSrc homology 2 domain containing adaptor protein B (204657_s_at), score: 0.6 SIGIRRsingle immunoglobulin and toll-interleukin 1 receptor (TIR) domain (52940_at), score: 0.65 SIN3BSIN3 homolog B, transcription regulator (yeast) (209352_s_at), score: 0.57 SIPA1signal-induced proliferation-associated 1 (204164_at), score: 0.57 SIRT6sirtuin (silent mating type information regulation 2 homolog) 6 (S. cerevisiae) (219613_s_at), score: 0.59 SLC25A13solute carrier family 25, member 13 (citrin) (203775_at), score: -0.72 SLC25A28solute carrier family 25, member 28 (221432_s_at), score: 0.57 SLC2A4RGSLC2A4 regulator (218494_s_at), score: 0.65 SLC30A9solute carrier family 30 (zinc transporter), member 9 (202614_at), score: -0.72 SLC33A1solute carrier family 33 (acetyl-CoA transporter), member 1 (203165_s_at), score: -0.7 SLC39A14solute carrier family 39 (zinc transporter), member 14 (212110_at), score: -0.61 SLC43A3solute carrier family 43, member 3 (213113_s_at), score: 0.66 SLC4A7solute carrier family 4, sodium bicarbonate cotransporter, member 7 (209884_s_at), score: -0.79 SLC5A5solute carrier family 5 (sodium iodide symporter), member 5 (211123_at), score: -0.68 SLC9A3R2solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 (209830_s_at), score: 0.57 SLMO2slowmo homolog 2 (Drosophila) (217851_s_at), score: -0.82 SMCHD1structural maintenance of chromosomes flexible hinge domain containing 1 (212569_at), score: -0.63 SMG6Smg-6 homolog, nonsense mediated mRNA decay factor (C. elegans) (214940_s_at), score: 0.76 SMN1survival of motor neuron 1, telomeric (203852_s_at), score: -0.65 SMYD5SMYD family member 5 (209516_at), score: 0.57 SNAP25synaptosomal-associated protein, 25kDa (202508_s_at), score: 0.58 SNX27sorting nexin family member 27 (221006_s_at), score: 0.64 SOAT1sterol O-acyltransferase 1 (221561_at), score: -0.59 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: 0.84 SORBS3sorbin and SH3 domain containing 3 (209253_at), score: 0.7 SOS2son of sevenless homolog 2 (Drosophila) (212870_at), score: -0.67 SP3Sp3 transcription factor (213168_at), score: -0.59 SPASTspastin (209748_at), score: -0.62 SPATA20spermatogenesis associated 20 (218164_at), score: 0.6 SPG20spastic paraplegia 20 (Troyer syndrome) (212526_at), score: -0.59 SPG7spastic paraplegia 7 (pure and complicated autosomal recessive) (202104_s_at), score: 0.64 SSBP3single stranded DNA binding protein 3 (217991_x_at), score: 0.68 ST20suppressor of tumorigenicity 20 (210242_x_at), score: -0.7 STAT6signal transducer and activator of transcription 6, interleukin-4 induced (201331_s_at), score: 0.61 STIM1stromal interaction molecule 1 (202764_at), score: 0.62 STRN4striatin, calmodulin binding protein 4 (217903_at), score: 0.67 SULT1B1sulfotransferase family, cytosolic, 1B, member 1 (207601_at), score: -0.77 SUPT6Hsuppressor of Ty 6 homolog (S. cerevisiae) (208831_x_at), score: 0.6 TAF2TAF2 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 150kDa (209523_at), score: -0.72 TAPBPTAP binding protein (tapasin) (208829_at), score: 0.65 TASP1taspase, threonine aspartase, 1 (219443_at), score: -0.61 TBCCD1TBCC domain containing 1 (206451_at), score: -0.63 TBCDtubulin folding cofactor D (211052_s_at), score: 0.68 TCEB1transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C) (202823_at), score: -0.65 TDGthymine-DNA glycosylase (203743_s_at), score: -0.61 TENC1tensin like C1 domain containing phosphatase (tensin 2) (212494_at), score: 0.71 TERF2telomeric repeat binding factor 2 (203611_at), score: 0.6 TGDSTDP-glucose 4,6-dehydratase (208249_s_at), score: -0.62 TIAF1TGFB1-induced anti-apoptotic factor 1 (202039_at), score: 0.76 TICAM2toll-like receptor adaptor molecule 2 (214658_at), score: -0.97 TLE3transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) (206472_s_at), score: 0.6 TLE4transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) (204872_at), score: -0.72 TM9SF3transmembrane 9 superfamily member 3 (217758_s_at), score: -0.59 TMED7transmembrane emp24 protein transport domain containing 7 (209404_s_at), score: -0.61 TMEM115transmembrane protein 115 (216267_s_at), score: 0.6 TMEM123transmembrane protein 123 (211967_at), score: -0.67 TMEM184Ctransmembrane protein 184C (219074_at), score: -0.58 TMEM39Btransmembrane protein 39B (218770_s_at), score: 0.63 TMSB4Ythymosin beta 4, Y-linked (206769_at), score: -0.64 TOMM20translocase of outer mitochondrial membrane 20 homolog (yeast) (200662_s_at), score: -0.6 TRIM21tripartite motif-containing 21 (204804_at), score: 0.72 TRIM24tripartite motif-containing 24 (213301_x_at), score: -0.57 TRIM3tripartite motif-containing 3 (204911_s_at), score: 0.57 TRMT2ATRM2 tRNA methyltransferase 2 homolog A (S. cerevisiae) (91617_at), score: -0.68 TSC2tuberous sclerosis 2 (215735_s_at), score: 0.71 TSKUtsukushin (218245_at), score: 0.74 TSPAN31tetraspanin 31 (203227_s_at), score: -0.61 TTC15tetratricopeptide repeat domain 15 (203122_at), score: 0.61 TUBA4Btubulin, alpha 4b (pseudogene) (207490_at), score: -0.72 TXLNAtaxilin alpha (212300_at), score: 0.59 TYRO3TYRO3 protein tyrosine kinase (211432_s_at), score: 0.63 UBA2ubiquitin-like modifier activating enzyme 2 (201177_s_at), score: -0.62 UBE2Zubiquitin-conjugating enzyme E2Z (217750_s_at), score: 0.6 UBTD1ubiquitin domain containing 1 (219172_at), score: 0.73 UFM1ubiquitin-fold modifier 1 (218050_at), score: -0.59 UGDHUDP-glucose dehydrogenase (203343_at), score: -0.64 ULK1unc-51-like kinase 1 (C. elegans) (209333_at), score: 0.65 USF2upstream transcription factor 2, c-fos interacting (202152_x_at), score: 0.74 USP12ubiquitin specific peptidase 12 (213327_s_at), score: -0.61 VAT1vesicle amine transport protein 1 homolog (T. californica) (208626_s_at), score: 0.66 VCPIP1valosin containing protein (p97)/p47 complex interacting protein 1 (219810_at), score: -0.6 VEGFBvascular endothelial growth factor B (203683_s_at), score: 1 VENTXP1VENT homeobox (Xenopus laevis) pseudogene 1 (216722_at), score: 0.61 VPS13Cvacuolar protein sorting 13 homolog C (S. cerevisiae) (218396_at), score: -0.7 VPS37Bvacuolar protein sorting 37 homolog B (S. cerevisiae) (221704_s_at), score: 0.85 VRK3vaccinia related kinase 3 (221998_s_at), score: 0.59 WBP2WW domain binding protein 2 (209117_at), score: 0.62 WDR4WD repeat domain 4 (221632_s_at), score: -0.73 WDR42AWD repeat domain 42A (202249_s_at), score: 0.62 WDR44WD repeat domain 44 (219297_at), score: -0.58 WDR6WD repeat domain 6 (217734_s_at), score: 0.69 WIZwidely interspaced zinc finger motifs (52005_at), score: 0.6 WNT2Bwingless-type MMTV integration site family, member 2B (206459_s_at), score: -0.59 WRNWerner syndrome (205667_at), score: -0.66 WWTR1WW domain containing transcription regulator 1 (202132_at), score: -0.64 XAB2XPA binding protein 2 (218110_at), score: 0.73 YTHDC2YTH domain containing 2 (213077_at), score: -0.78 YTHDF3YTH domain family, member 3 (221749_at), score: -0.67 ZBTB10zinc finger and BTB domain containing 10 (219312_s_at), score: -0.65 ZC3H14zinc finger CCCH-type containing 14 (213064_at), score: -0.58 ZC3H3zinc finger CCCH-type containing 3 (213445_at), score: 0.63 ZCCHC10zinc finger, CCHC domain containing 10 (221193_s_at), score: -0.84 ZDHHC17zinc finger, DHHC-type containing 17 (212982_at), score: -0.61 ZFAND1zinc finger, AN1-type domain 1 (218919_at), score: -0.64 ZFHX3zinc finger homeobox 3 (208033_s_at), score: 0.63 ZFP36L1zinc finger protein 36, C3H type-like 1 (211965_at), score: 0.66 ZFYVE16zinc finger, FYVE domain containing 16 (203651_at), score: -0.67 ZFYVE26zinc finger, FYVE domain containing 26 (37943_at), score: -0.61 ZNF248zinc finger protein 248 (213269_at), score: -0.61 ZNF580zinc finger protein 580 (220748_s_at), score: 0.7 ZNF706zinc finger protein 706 (218059_at), score: 0.58 ZYXzyxin (200808_s_at), score: 0.57
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690272.cel | 7 | 5 | HGPS | hgu133a | HGPS | AG11498 |
| E-TABM-263-raw-cel-1515486251.cel | 31 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485991.cel | 18 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690304.cel | 8 | 5 | HGPS | hgu133a | none | GMO8398C |
| 10358_WBS.CEL | 1 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| 10590_WBS.CEL | 2 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486411.cel | 39 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486071.cel | 22 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |