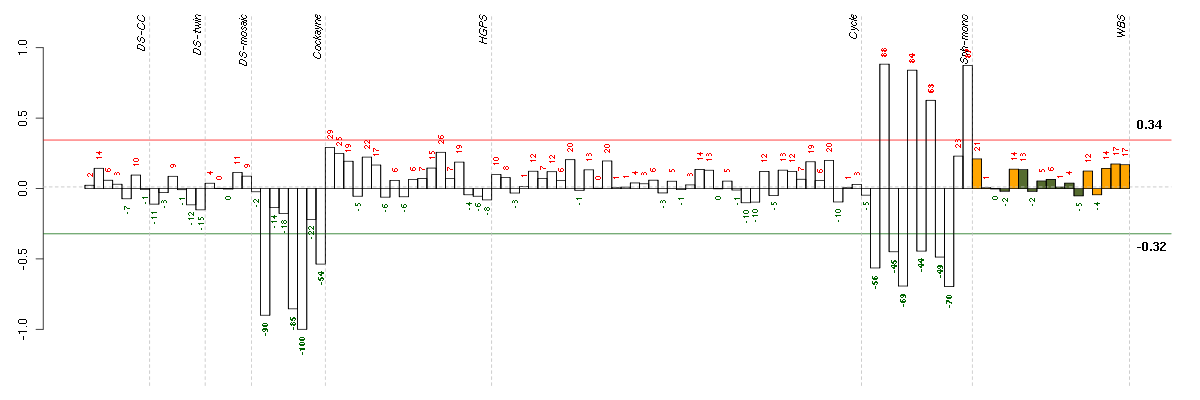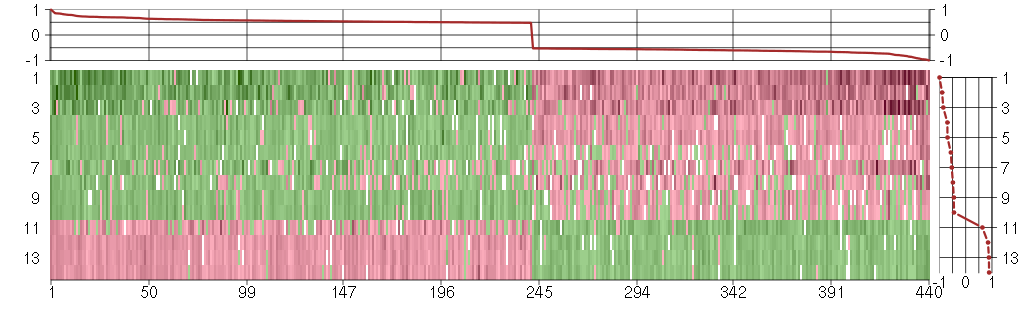

Under-expression is coded with green,
over-expression with red color.



| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 00150 | 5.470e-03 | 1.079 | 7 | 21 | Androgen and estrogen metabolism |
| 00980 | 2.846e-02 | 1.9 | 8 | 37 | Metabolism of xenobiotics by cytochrome P450 |
ABCB1ATP-binding cassette, sub-family B (MDR/TAP), member 1 (209993_at), score: -0.89 ABCB4ATP-binding cassette, sub-family B (MDR/TAP), member 4 (207819_s_at), score: -0.72 ABCC10ATP-binding cassette, sub-family C (CFTR/MRP), member 10 (213485_s_at), score: 0.55 ACTA2actin, alpha 2, smooth muscle, aorta (200974_at), score: 0.5 ADCY7adenylate cyclase 7 (203741_s_at), score: 0.55 ADORA2Badenosine A2b receptor (205891_at), score: -0.65 AEBP1AE binding protein 1 (201792_at), score: 0.62 AGPSalkylglycerone phosphate synthase (205401_at), score: -0.69 AIM2absent in melanoma 2 (206513_at), score: -0.55 AKAP8LA kinase (PRKA) anchor protein 8-like (218064_s_at), score: -0.54 AKR1B10aldo-keto reductase family 1, member B10 (aldose reductase) (206561_s_at), score: -0.94 AKR1C3aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II) (209160_at), score: -0.57 ALDH3A1aldehyde dehydrogenase 3 family, memberA1 (205623_at), score: -0.53 ANGangiogenin, ribonuclease, RNase A family, 5 (213397_x_at), score: 0.48 ANK3ankyrin 3, node of Ranvier (ankyrin G) (206385_s_at), score: -0.59 ANPEPalanyl (membrane) aminopeptidase (202888_s_at), score: 0.56 APH1Banterior pharynx defective 1 homolog B (C. elegans) (221036_s_at), score: 0.68 APOBEC3Gapolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3G (204205_at), score: -0.64 APOL3apolipoprotein L, 3 (221087_s_at), score: -0.61 APPL2adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 (218218_at), score: 0.5 AQP3aquaporin 3 (Gill blood group) (39248_at), score: -0.55 ARHGAP5Rho GTPase activating protein 5 (217936_at), score: 0.57 ARMCX1armadillo repeat containing, X-linked 1 (218694_at), score: 0.58 ARMCX3armadillo repeat containing, X-linked 3 (217858_s_at), score: 0.53 ARSJarylsulfatase family, member J (219973_at), score: 0.67 ASCC3activating signal cointegrator 1 complex subunit 3 (212815_at), score: 0.5 ASPNasporin (219087_at), score: 0.49 ATP1A1ATPase, Na+/K+ transporting, alpha 1 polypeptide (220948_s_at), score: -0.6 ATP2B4ATPase, Ca++ transporting, plasma membrane 4 (212135_s_at), score: 0.52 AUP1ancient ubiquitous protein 1 (220525_s_at), score: -0.54 BAMBIBMP and activin membrane-bound inhibitor homolog (Xenopus laevis) (203304_at), score: -0.53 BAT1HLA-B associated transcript 1 (200041_s_at), score: -0.56 BAZ2Bbromodomain adjacent to zinc finger domain, 2B (203080_s_at), score: 0.53 BIRC3baculoviral IAP repeat-containing 3 (210538_s_at), score: -0.56 BMP7bone morphogenetic protein 7 (209590_at), score: -0.54 BRF2BRF2, subunit of RNA polymerase III transcription initiation factor, BRF1-like (218955_at), score: -0.58 C10orf116chromosome 10 open reading frame 116 (203571_s_at), score: 0.68 C12orf47chromosome 12 open reading frame 47 (64432_at), score: -0.59 C14orf169chromosome 14 open reading frame 169 (219526_at), score: 0.52 C15orf29chromosome 15 open reading frame 29 (218791_s_at), score: 0.53 C1orf2chromosome 1 open reading frame 2 (203550_s_at), score: 0.49 C20orf111chromosome 20 open reading frame 111 (209020_at), score: -0.61 C20orf149chromosome 20 open reading frame 149 (218010_x_at), score: 0.53 C20orf4chromosome 20 open reading frame 4 (218089_at), score: -0.53 C21orf59chromosome 21 open reading frame 59 (218123_at), score: -0.55 C21orf66chromosome 21 open reading frame 66 (221158_at), score: -0.56 C3orf64chromosome 3 open reading frame 64 (221935_s_at), score: 0.71 C4orf30chromosome 4 open reading frame 30 (219717_at), score: -0.56 C5orf30chromosome 5 open reading frame 30 (221823_at), score: 0.62 C5orf4chromosome 5 open reading frame 4 (48031_r_at), score: 0.58 C6orf120chromosome 6 open reading frame 120 (221786_at), score: 0.48 C7orf64chromosome 7 open reading frame 64 (221596_s_at), score: -0.56 C9orf125chromosome 9 open reading frame 125 (213386_at), score: 0.5 CALML4calmodulin-like 4 (64408_s_at), score: -0.62 CCDC59coiled-coil domain containing 59 (218936_s_at), score: -0.58 CCNG1cyclin G1 (208796_s_at), score: 0.63 CCNL2cyclin L2 (221427_s_at), score: -0.56 CCRL1chemokine (C-C motif) receptor-like 1 (220351_at), score: 0.59 CD24CD24 molecule (209771_x_at), score: -0.83 CD248CD248 molecule, endosialin (219025_at), score: 0.79 CD81CD81 molecule (200675_at), score: 0.57 CDC25Ccell division cycle 25 homolog C (S. pombe) (205167_s_at), score: -0.54 CDK5RAP1CDK5 regulatory subunit associated protein 1 (218315_s_at), score: -0.6 CDKN1Acyclin-dependent kinase inhibitor 1A (p21, Cip1) (202284_s_at), score: 0.79 CDKN2Acyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4) (209644_x_at), score: 0.49 CELSR1cadherin, EGF LAG seven-pass G-type receptor 1 (flamingo homolog, Drosophila) (41660_at), score: -0.61 CELSR3cadherin, EGF LAG seven-pass G-type receptor 3 (flamingo homolog, Drosophila) (40020_at), score: -0.66 CEP170centrosomal protein 170kDa (212746_s_at), score: 0.55 CFHcomplement factor H (213800_at), score: 0.52 CHKAcholine kinase alpha (204233_s_at), score: -0.63 CHMP1Achromatin modifying protein 1A (201933_at), score: -0.63 CHN1chimerin (chimaerin) 1 (212624_s_at), score: 0.49 CLEC11AC-type lectin domain family 11, member A (211709_s_at), score: 0.62 CNTNAP1contactin associated protein 1 (219400_at), score: 0.48 COL11A1collagen, type XI, alpha 1 (37892_at), score: 0.7 COL3A1collagen, type III, alpha 1 (215076_s_at), score: 0.67 CRABP2cellular retinoic acid binding protein 2 (202575_at), score: 0.57 CRATcarnitine acetyltransferase (209522_s_at), score: 0.52 CREB3L1cAMP responsive element binding protein 3-like 1 (213059_at), score: 0.49 CRTAPcartilage associated protein (201380_at), score: 0.58 CRYABcrystallin, alpha B (209283_at), score: 0.57 CTNNBL1catenin, beta like 1 (221021_s_at), score: -0.67 CTSBcathepsin B (213274_s_at), score: 0.49 CTSHcathepsin H (202295_s_at), score: -0.65 CTSKcathepsin K (202450_s_at), score: 0.7 CYB5R3cytochrome b5 reductase 3 (201885_s_at), score: 0.5 CYBRD1cytochrome b reductase 1 (217889_s_at), score: 0.5 DBF4DBF4 homolog (S. cerevisiae) (204244_s_at), score: -0.58 DCHS1dachsous 1 (Drosophila) (222101_s_at), score: 0.51 DCNdecorin (211896_s_at), score: 0.7 DDAH1dimethylarginine dimethylaminohydrolase 1 (209094_at), score: 0.7 DDX3YDEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked (205000_at), score: 0.5 DERL1Der1-like domain family, member 1 (219402_s_at), score: -0.54 DGKDdiacylglycerol kinase, delta 130kDa (208072_s_at), score: -0.96 DHX38DEAH (Asp-Glu-Ala-His) box polypeptide 38 (209178_at), score: -0.62 DIO2deiodinase, iodothyronine, type II (203699_s_at), score: -0.78 DKFZP586H2123regeneration associated muscle protease (213661_at), score: 0.58 DNAJC7DnaJ (Hsp40) homolog, subfamily C, member 7 (202416_at), score: -0.69 DNASE1L1deoxyribonuclease I-like 1 (203912_s_at), score: 0.54 DRAP1DR1-associated protein 1 (negative cofactor 2 alpha) (203258_at), score: -0.61 DRG1developmentally regulated GTP binding protein 1 (202810_at), score: -0.55 DSEdermatan sulfate epimerase (218854_at), score: 0.6 DSPdesmoplakin (200606_at), score: 0.57 DUSP14dual specificity phosphatase 14 (203367_at), score: 0.57 EDNRAendothelin receptor type A (204464_s_at), score: 0.51 EIF3Deukaryotic translation initiation factor 3, subunit D (200005_at), score: -0.53 EIF4A1eukaryotic translation initiation factor 4A, isoform 1 (211787_s_at), score: -0.55 EIF6eukaryotic translation initiation factor 6 (210213_s_at), score: -0.62 ELOVL4elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 4 (219532_at), score: 0.54 EMP1epithelial membrane protein 1 (201325_s_at), score: 0.58 ENPP2ectonucleotide pyrophosphatase/phosphodiesterase 2 (209392_at), score: 0.63 EPHB2EPH receptor B2 (209589_s_at), score: -0.68 EPHX1epoxide hydrolase 1, microsomal (xenobiotic) (202017_at), score: -0.71 EVI1ecotropic viral integration site 1 (221884_at), score: -0.54 EWSR1Ewing sarcoma breakpoint region 1 (209214_s_at), score: -0.67 FAF2Fas associated factor family member 2 (212106_at), score: -0.57 FAM105Afamily with sequence similarity 105, member A (219694_at), score: -0.79 FAM169Afamily with sequence similarity 169, member A (213954_at), score: -0.57 FAM174Bfamily with sequence similarity 174, member B (51158_at), score: 0.49 FAPfibroblast activation protein, alpha (209955_s_at), score: 0.5 FARP1FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) (201911_s_at), score: 0.59 FAT1FAT tumor suppressor homolog 1 (Drosophila) (201579_at), score: 0.75 FBN2fibrillin 2 (203184_at), score: 0.68 FGF2fibroblast growth factor 2 (basic) (204421_s_at), score: 0.51 FJX1four jointed box 1 (Drosophila) (219522_at), score: 0.53 FKBP11FK506 binding protein 11, 19 kDa (219118_at), score: 0.57 FKBP4FK506 binding protein 4, 59kDa (200895_s_at), score: -0.53 FKBP9FK506 binding protein 9, 63 kDa (212169_at), score: 0.52 FLJ10357hypothetical protein FLJ10357 (58780_s_at), score: 0.67 FLJ12529pre-mRNA cleavage factor I, 59 kDa subunit (217866_at), score: -0.55 FLRT2fibronectin leucine rich transmembrane protein 2 (204359_at), score: 0.52 FOXG1forkhead box G1 (206018_at), score: -0.8 FOXL2forkhead box L2 (220102_at), score: -0.69 GABREgamma-aminobutyric acid (GABA) A receptor, epsilon (204537_s_at), score: -0.57 GAGE4G antigen 4 (207086_x_at), score: -0.85 GAGE6G antigen 6 (208155_x_at), score: -0.82 GAL3ST4galactose-3-O-sulfotransferase 4 (219815_at), score: 0.52 GALCgalactosylceramidase (204417_at), score: 0.69 GALNT6UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 6 (GalNAc-T6) (219956_at), score: 0.53 GAS1growth arrest-specific 1 (204457_s_at), score: 0.67 GAS6growth arrest-specific 6 (202177_at), score: 0.49 GATA3GATA binding protein 3 (209604_s_at), score: -0.76 GCLCglutamate-cysteine ligase, catalytic subunit (202923_s_at), score: -0.7 GHRgrowth hormone receptor (205498_at), score: 0.48 GIPC2GIPC PDZ domain containing family, member 2 (219970_at), score: 0.48 GLSglutaminase (203159_at), score: 0.71 GLT25D2glycosyltransferase 25 domain containing 2 (209883_at), score: 0.51 GNL3guanine nucleotide binding protein-like 3 (nucleolar) (217850_at), score: -0.67 GPRC5BG protein-coupled receptor, family C, group 5, member B (203632_s_at), score: -0.69 GRAMD3GRAM domain containing 3 (218706_s_at), score: 0.55 GREM1gremlin 1, cysteine knot superfamily, homolog (Xenopus laevis) (218469_at), score: 1 GRIK2glutamate receptor, ionotropic, kainate 2 (213845_at), score: 0.56 GSRglutathione reductase (205770_at), score: -0.59 GSTP1glutathione S-transferase pi 1 (200824_at), score: -0.53 HAS2hyaluronan synthase 2 (206432_at), score: 0.56 HEPHhephaestin (203903_s_at), score: 0.58 HERC5hect domain and RLD 5 (219863_at), score: -0.81 HEY1hairy/enhancer-of-split related with YRPW motif 1 (44783_s_at), score: -0.58 HGSNATheparan-alpha-glucosaminide N-acetyltransferase (218017_s_at), score: 0.69 HIST1H2BKhistone cluster 1, H2bk (209806_at), score: 0.51 HIVEP2human immunodeficiency virus type I enhancer binding protein 2 (212642_s_at), score: 0.5 HLA-Bmajor histocompatibility complex, class I, B (211911_x_at), score: -0.7 HLA-Cmajor histocompatibility complex, class I, C (211799_x_at), score: -0.6 HLA-DRB1major histocompatibility complex, class II, DR beta 1 (209312_x_at), score: -0.59 HLA-Fmajor histocompatibility complex, class I, F (204806_x_at), score: -0.65 HOXA10homeobox A10 (213150_at), score: 0.63 HOXA11homeobox A11 (213823_at), score: 0.49 HRASLSHRAS-like suppressor (219983_at), score: -0.55 HS3ST3A1heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1 (219985_at), score: 0.51 HSD17B12hydroxysteroid (17-beta) dehydrogenase 12 (217869_at), score: 0.7 HSP90AB1heat shock protein 90kDa alpha (cytosolic), class B member 1 (214359_s_at), score: -0.65 HSPB6heat shock protein, alpha-crystallin-related, B6 (214767_s_at), score: 0.48 HYPKHuntingtin interacting protein K (218680_x_at), score: -0.53 IDH3Bisocitrate dehydrogenase 3 (NAD+) beta (210014_x_at), score: -0.54 IFI44interferon-induced protein 44 (214453_s_at), score: -0.54 IGF2BP3insulin-like growth factor 2 mRNA binding protein 3 (203819_s_at), score: -0.62 IGFBP4insulin-like growth factor binding protein 4 (201508_at), score: 0.54 IGSF3immunoglobulin superfamily, member 3 (202421_at), score: -0.91 IKIK cytokine, down-regulator of HLA II (200066_at), score: -0.73 IL1R1interleukin 1 receptor, type I (202948_at), score: 0.5 INAinternexin neuronal intermediate filament protein, alpha (204465_s_at), score: 0.52 INPP4Binositol polyphosphate-4-phosphatase, type II, 105kDa (205376_at), score: 0.49 IPWimprinted in Prader-Willi syndrome (non-protein coding) (221974_at), score: 0.56 IQWD1IQ motif and WD repeats 1 (217908_s_at), score: 0.51 IRS2insulin receptor substrate 2 (209185_s_at), score: 0.57 ITGA5integrin, alpha 5 (fibronectin receptor, alpha polypeptide) (201389_at), score: 0.61 KAT2BK(lysine) acetyltransferase 2B (203845_at), score: 0.54 KCNK2potassium channel, subfamily K, member 2 (210261_at), score: 0.55 KIAA0802KIAA0802 (213358_at), score: 0.55 KIAA0895KIAA0895 (213424_at), score: -0.6 KIAA1128KIAA1128 (209379_s_at), score: 0.53 KIAA1797KIAA1797 (218503_at), score: 0.48 KIF3Bkinesin family member 3B (203943_at), score: -0.63 KIFC1kinesin family member C1 (209680_s_at), score: -0.55 KPNA1karyopherin alpha 1 (importin alpha 5) (202055_at), score: 0.52 KRT34keratin 34 (206969_at), score: 0.54 KYNUkynureninase (L-kynurenine hydrolase) (217388_s_at), score: -0.97 LAMB3laminin, beta 3 (209270_at), score: -0.62 LBHlimb bud and heart development homolog (mouse) (221011_s_at), score: 0.55 LDOC1leucine zipper, down-regulated in cancer 1 (204454_at), score: 0.5 LEF1lymphoid enhancer-binding factor 1 (221558_s_at), score: -1 LGMNlegumain (201212_at), score: 0.59 LHFPlipoma HMGIC fusion partner (218656_s_at), score: 0.55 LIPAlipase A, lysosomal acid, cholesterol esterase (201847_at), score: 0.64 LMO7LIM domain 7 (202674_s_at), score: 0.62 LOC100129250similar to hCG1811779 (78383_at), score: 0.48 LOC100130886hypothetical protein LOC100130886 (220486_x_at), score: -0.57 LOC388796hypothetical LOC388796 (65588_at), score: -0.63 LOC728855hypothetical LOC728855 (222001_x_at), score: -0.88 LPPR4plasticity related gene 1 (213496_at), score: 0.56 LRP10low density lipoprotein receptor-related protein 10 (201412_at), score: 0.54 LRRC2leucine rich repeat containing 2 (219949_at), score: 0.79 LSG1large subunit GTPase 1 homolog (S. cerevisiae) (221536_s_at), score: -0.64 LTBP2latent transforming growth factor beta binding protein 2 (204682_at), score: 0.85 LUMlumican (201744_s_at), score: 0.85 LYPD1LY6/PLAUR domain containing 1 (212909_at), score: -0.54 MAFBv-maf musculoaponeurotic fibrosarcoma oncogene homolog B (avian) (218559_s_at), score: -0.56 MAGOHmago-nashi homolog, proliferation-associated (Drosophila) (210092_at), score: -0.73 MAN2A1mannosidase, alpha, class 2A, member 1 (205105_at), score: 0.69 MANSC1MANSC domain containing 1 (220945_x_at), score: 0.51 MARCH2membrane-associated ring finger (C3HC4) 2 (210075_at), score: 0.61 MFAP5microfibrillar associated protein 5 (213764_s_at), score: 0.57 MLF1myeloid leukemia factor 1 (204784_s_at), score: -0.64 MMEmembrane metallo-endopeptidase (203434_s_at), score: 0.56 MMP1matrix metallopeptidase 1 (interstitial collagenase) (204475_at), score: 0.53 MOXD1monooxygenase, DBH-like 1 (209708_at), score: 0.81 MRC2mannose receptor, C type 2 (37408_at), score: 0.69 MRPL22mitochondrial ribosomal protein L22 (218339_at), score: -0.57 MRPL28mitochondrial ribosomal protein L28 (204599_s_at), score: -0.59 MRPL9mitochondrial ribosomal protein L9 (211594_s_at), score: -0.56 MTF2metal response element binding transcription factor 2 (203345_s_at), score: -0.55 MTUS1mitochondrial tumor suppressor 1 (212096_s_at), score: -0.73 MXRA5matrix-remodelling associated 5 (209596_at), score: 0.71 MXRA8matrix-remodelling associated 8 (213422_s_at), score: 0.71 MYO1Bmyosin IB (212364_at), score: 0.73 MYO1Dmyosin ID (212338_at), score: 0.49 MYST4MYST histone acetyltransferase (monocytic leukemia) 4 (212462_at), score: 0.48 NASPnuclear autoantigenic sperm protein (histone-binding) (201970_s_at), score: -0.62 NBEAneurobeachin (221207_s_at), score: 0.53 NDNnecdin homolog (mouse) (209550_at), score: 0.69 NDUFS3NADH dehydrogenase (ubiquinone) Fe-S protein 3, 30kDa (NADH-coenzyme Q reductase) (201740_at), score: -0.57 NEDD4Lneural precursor cell expressed, developmentally down-regulated 4-like (212445_s_at), score: -0.55 NEFLneurofilament, light polypeptide (221805_at), score: -0.6 NEK7NIMA (never in mitosis gene a)-related kinase 7 (212530_at), score: 0.69 NFASCneurofascin homolog (chicken) (213438_at), score: 0.62 NGDNneuroguidin, EIF4E binding protein (213794_s_at), score: -0.65 NIP7nuclear import 7 homolog (S. cerevisiae) (219031_s_at), score: -0.64 NLGN1neuroligin 1 (205893_at), score: 0.49 NMUneuromedin U (206023_at), score: -0.64 NOC2Lnucleolar complex associated 2 homolog (S. cerevisiae) (202115_s_at), score: -0.55 NOL12nucleolar protein 12 (219324_at), score: -0.67 NONOnon-POU domain containing, octamer-binding (208698_s_at), score: -0.63 NR2F1nuclear receptor subfamily 2, group F, member 1 (209505_at), score: -0.55 NRGNneurogranin (protein kinase C substrate, RC3) (204081_at), score: -0.58 NRP1neuropilin 1 (212298_at), score: 0.51 NSFL1CNSFL1 (p97) cofactor (p47) (217831_s_at), score: -0.58 NT5E5'-nucleotidase, ecto (CD73) (203939_at), score: 0.63 OASL2'-5'-oligoadenylate synthetase-like (210797_s_at), score: -0.85 ODZ4odz, odd Oz/ten-m homolog 4 (Drosophila) (213273_at), score: 0.49 OLFML3olfactomedin-like 3 (218162_at), score: 0.64 OPTNoptineurin (202073_at), score: 0.52 OXTRoxytocin receptor (206825_at), score: 0.55 P4HA1prolyl 4-hydroxylase, alpha polypeptide I (207543_s_at), score: 0.58 PBX1pre-B-cell leukemia homeobox 1 (212148_at), score: 0.73 PCLOpiccolo (presynaptic cytomatrix protein) (213558_at), score: -0.96 PCNXpecanex homolog (Drosophila) (213173_at), score: 0.48 PCTK2PCTAIRE protein kinase 2 (221918_at), score: 0.58 PDGFCplatelet derived growth factor C (218718_at), score: 0.69 PDGFRAplatelet-derived growth factor receptor, alpha polypeptide (203131_at), score: 0.62 PDZRN3PDZ domain containing ring finger 3 (212915_at), score: 0.59 PEX5peroxisomal biogenesis factor 5 (203244_at), score: -0.59 PFDN2prefoldin subunit 2 (218336_at), score: -0.62 PFDN6prefoldin subunit 6 (222029_x_at), score: -0.56 PGAP1post-GPI attachment to proteins 1 (213469_at), score: 0.6 PHF11PHD finger protein 11 (221816_s_at), score: 0.48 PID1phosphotyrosine interaction domain containing 1 (219093_at), score: 0.6 PILRBpaired immunoglobin-like type 2 receptor beta (220954_s_at), score: -0.63 PIRpirin (iron-binding nuclear protein) (207469_s_at), score: -0.6 PJA2praja ring finger 2 (201133_s_at), score: 0.5 PKD2polycystic kidney disease 2 (autosomal dominant) (203688_at), score: 0.57 PLAGL1pleiomorphic adenoma gene-like 1 (209318_x_at), score: 0.48 PLCB1phospholipase C, beta 1 (phosphoinositide-specific) (213222_at), score: 0.66 PLK2polo-like kinase 2 (Drosophila) (201939_at), score: -0.65 POLR3Gpolymerase (RNA) III (DNA directed) polypeptide G (32kD) (206653_at), score: -0.6 POSTNperiostin, osteoblast specific factor (210809_s_at), score: 0.56 PPIEpeptidylprolyl isomerase E (cyclophilin E) (210502_s_at), score: -0.6 PPIHpeptidylprolyl isomerase H (cyclophilin H) (204228_at), score: -0.7 PPM2Cprotein phosphatase 2C, magnesium-dependent, catalytic subunit (218273_s_at), score: -0.55 PPP1R3Dprotein phosphatase 1, regulatory (inhibitor) subunit 3D (204554_at), score: 0.54 PPP2R2Bprotein phosphatase 2 (formerly 2A), regulatory subunit B, beta isoform (213849_s_at), score: -0.57 PPP3CBprotein phosphatase 3 (formerly 2B), catalytic subunit, beta isoform (209817_at), score: 0.58 PQBP1polyglutamine binding protein 1 (214527_s_at), score: -0.72 PRKCSHprotein kinase C substrate 80K-H (214080_x_at), score: 0.51 PRR16proline rich 16 (220014_at), score: 0.62 PRRX1paired related homeobox 1 (205991_s_at), score: 0.63 PSG2pregnancy specific beta-1-glycoprotein 2 (208134_x_at), score: 0.54 PSG5pregnancy specific beta-1-glycoprotein 5 (204830_x_at), score: 0.53 PSG9pregnancy specific beta-1-glycoprotein 9 (209594_x_at), score: 0.48 PSMB1proteasome (prosome, macropain) subunit, beta type, 1 (200876_s_at), score: -0.54 PSMB8proteasome (prosome, macropain) subunit, beta type, 8 (large multifunctional peptidase 7) (209040_s_at), score: -0.72 PSMC3proteasome (prosome, macropain) 26S subunit, ATPase, 3 (201267_s_at), score: -0.6 PSMD13proteasome (prosome, macropain) 26S subunit, non-ATPase, 13 (201232_s_at), score: -0.59 PSMD4proteasome (prosome, macropain) 26S subunit, non-ATPase, 4 (211609_x_at), score: -0.61 PSTPIP2proline-serine-threonine phosphatase interacting protein 2 (219938_s_at), score: 0.63 PTGS1prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) (215813_s_at), score: 0.54 PTGS2prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) (204748_at), score: 0.5 PTPRDprotein tyrosine phosphatase, receptor type, D (214043_at), score: 0.53 QPCTglutaminyl-peptide cyclotransferase (205174_s_at), score: 0.72 R3HCC1R3H domain and coiled-coil containing 1 (212866_at), score: -0.54 R3HDM2R3H domain containing 2 (203831_at), score: 0.55 RAB27ARAB27A, member RAS oncogene family (210951_x_at), score: -0.55 RAB7L1RAB7, member RAS oncogene family-like 1 (218700_s_at), score: -0.53 RALYRNA binding protein, autoantigenic (hnRNP-associated with lethal yellow homolog (mouse)) (201271_s_at), score: -0.61 RARRES3retinoic acid receptor responder (tazarotene induced) 3 (204070_at), score: -0.66 RECKreversion-inducing-cysteine-rich protein with kazal motifs (205407_at), score: 0.84 RGP1RGP1 retrograde golgi transport homolog (S. cerevisiae) (203169_at), score: -0.72 RGS4regulator of G-protein signaling 4 (204337_at), score: 0.62 RNF121ring finger protein 121 (219021_at), score: -0.57 ROCK1Rho-associated, coiled-coil containing protein kinase 1 (213044_at), score: 0.56 RPP14ribonuclease P/MRP 14kDa subunit (204245_s_at), score: -0.69 RPS20ribosomal protein S20 (216247_at), score: 0.51 RPS27Lribosomal protein S27-like (218007_s_at), score: 0.51 RPS3ribosomal protein S3 (208692_at), score: -0.55 RPS4Xribosomal protein S4, X-linked (200933_x_at), score: -0.54 RPS6KA1ribosomal protein S6 kinase, 90kDa, polypeptide 1 (203379_at), score: -0.53 RSL1D1ribosomal L1 domain containing 1 (212018_s_at), score: -0.66 RUNX1runt-related transcription factor 1 (209360_s_at), score: 0.55 S100A4S100 calcium binding protein A4 (203186_s_at), score: 0.57 SAMD4Asterile alpha motif domain containing 4A (212845_at), score: 0.73 SC65synaptonemal complex protein SC65 (204078_at), score: 0.57 SCRIBscribbled homolog (Drosophila) (212556_at), score: 0.52 SDHBsuccinate dehydrogenase complex, subunit B, iron sulfur (Ip) (202675_at), score: -0.55 SEMA3Fsema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F (35666_at), score: 0.56 SERPINB7serpin peptidase inhibitor, clade B (ovalbumin), member 7 (206421_s_at), score: 0.74 SERPINE2serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 (212190_at), score: 0.54 SERPINF1serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 (202283_at), score: 0.59 SGCDsarcoglycan, delta (35kDa dystrophin-associated glycoprotein) (213543_at), score: 0.48 SH3YL1SH3 domain containing, Ysc84-like 1 (S. cerevisiae) (204019_s_at), score: 0.55 SHFM1split hand/foot malformation (ectrodactyly) type 1 (202276_at), score: -0.67 SIM2single-minded homolog 2 (Drosophila) (206558_at), score: -0.6 SLC12A8solute carrier family 12 (potassium/chloride transporters), member 8 (219874_at), score: -0.7 SLC25A13solute carrier family 25, member 13 (citrin) (203775_at), score: -0.66 SLC25A37solute carrier family 25, member 37 (218136_s_at), score: -0.6 SLC25A5solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 (200657_at), score: -0.53 SLC2A10solute carrier family 2 (facilitated glucose transporter), member 10 (221024_s_at), score: 0.49 SLC30A1solute carrier family 30 (zinc transporter), member 1 (212907_at), score: 0.7 SLC39A4solute carrier family 39 (zinc transporter), member 4 (219215_s_at), score: 0.54 SLC4A4solute carrier family 4, sodium bicarbonate cotransporter, member 4 (203908_at), score: 0.53 SLC7A11solute carrier family 7, (cationic amino acid transporter, y+ system) member 11 (217678_at), score: -0.7 SMA5glucuronidase, beta pseudogene (215043_s_at), score: -0.59 SMAD5SMAD family member 5 (205187_at), score: -0.53 SMCR7LSmith-Magenis syndrome chromosome region, candidate 7-like (221516_s_at), score: -0.62 SMG1SMG1 homolog, phosphatidylinositol 3-kinase-related kinase (C. elegans) (210057_at), score: -0.62 SMPD1sphingomyelin phosphodiesterase 1, acid lysosomal (209420_s_at), score: 0.52 SNF8SNF8, ESCRT-II complex subunit, homolog (S. cerevisiae) (218391_at), score: -0.54 SNRPA1small nuclear ribonucleoprotein polypeptide A' (216977_x_at), score: -0.54 SNRPBsmall nuclear ribonucleoprotein polypeptides B and B1 (213175_s_at), score: -0.55 SOS2son of sevenless homolog 2 (Drosophila) (212870_at), score: 0.53 SPHARS-phase response (cyclin-related) (206272_at), score: 0.63 SQSTM1sequestosome 1 (201471_s_at), score: -0.55 SSPNsarcospan (Kras oncogene-associated gene) (204963_at), score: 0.61 SSR3signal sequence receptor, gamma (translocon-associated protein gamma) (217790_s_at), score: 0.6 SSRP1structure specific recognition protein 1 (200956_s_at), score: -0.65 ST3GAL6ST3 beta-galactoside alpha-2,3-sialyltransferase 6 (210942_s_at), score: -0.59 STAM2signal transducing adaptor molecule (SH3 domain and ITAM motif) 2 (209649_at), score: 0.49 STAU2staufen, RNA binding protein, homolog 2 (Drosophila) (204226_at), score: 0.48 STIP1stress-induced-phosphoprotein 1 (212009_s_at), score: -0.58 STOML2stomatin (EPB72)-like 2 (215416_s_at), score: -0.58 STSsteroid sulfatase (microsomal), isozyme S (203767_s_at), score: 0.61 STX2syntaxin 2 (213434_at), score: 0.58 STXBP6syntaxin binding protein 6 (amisyn) (220994_s_at), score: 0.57 SULF1sulfatase 1 (212353_at), score: 0.65 SULT1A1sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 (203615_x_at), score: -0.54 SULT1A2sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2 (207122_x_at), score: -0.56 SYNGR1synaptogyrin 1 (210613_s_at), score: 0.56 SYTL2synaptotagmin-like 2 (220613_s_at), score: -0.53 TAX1BP3Tax1 (human T-cell leukemia virus type I) binding protein 3 (215464_s_at), score: 0.55 TBC1D3TBC1 domain family, member 3 (209403_at), score: -0.63 TBC1D8TBC1 domain family, member 8 (with GRAM domain) (204526_s_at), score: -0.64 TBC1D9TBC1 domain family, member 9 (with GRAM domain) (212956_at), score: 0.58 TBXA2Rthromboxane A2 receptor (336_at), score: 0.64 TCEAL2transcription elongation factor A (SII)-like 2 (211276_at), score: -0.67 TCF12transcription factor 12 (208986_at), score: 0.5 TCF7L2transcription factor 7-like 2 (T-cell specific, HMG-box) (212761_at), score: 0.51 TEKTEK tyrosine kinase, endothelial (206702_at), score: 0.51 TEStestis derived transcript (3 LIM domains) (202720_at), score: 0.54 TESK1testis-specific kinase 1 (204106_at), score: 0.54 TGFB2transforming growth factor, beta 2 (220407_s_at), score: -0.62 THBS2thrombospondin 2 (203083_at), score: 0.48 TIMP2TIMP metallopeptidase inhibitor 2 (203167_at), score: 0.81 TKTtransketolase (208699_x_at), score: -0.63 TMEFF1transmembrane protein with EGF-like and two follistatin-like domains 1 (205122_at), score: 0.51 TMEM111transmembrane protein 111 (217882_at), score: -0.59 TMEM168transmembrane protein 168 (218962_s_at), score: 0.48 TMEM2transmembrane protein 2 (218113_at), score: 0.53 TMEM204transmembrane protein 204 (219315_s_at), score: 0.6 TMEM30Atransmembrane protein 30A (217743_s_at), score: 0.51 TMEM47transmembrane protein 47 (209656_s_at), score: 0.76 TMEM80transmembrane protein 80 (65630_at), score: -0.57 TMOD1tropomodulin 1 (203661_s_at), score: -0.57 TNS1tensin 1 (221748_s_at), score: 0.93 TOB1transducer of ERBB2, 1 (202704_at), score: 0.69 TPD52tumor protein D52 (201690_s_at), score: -0.67 TRAFD1TRAF-type zinc finger domain containing 1 (35254_at), score: -0.56 TRIM16tripartite motif-containing 16 (204341_at), score: -0.6 TRMUtRNA 5-methylaminomethyl-2-thiouridylate methyltransferase (213634_s_at), score: -0.54 TRPC3transient receptor potential cation channel, subfamily C, member 3 (210814_at), score: -0.55 TSPAN13tetraspanin 13 (217979_at), score: 0.78 TSPAN6tetraspanin 6 (209108_at), score: 0.5 TSPYL1TSPY-like 1 (221493_at), score: 0.58 TSPYL5TSPY-like 5 (213122_at), score: 0.84 TUBB6tubulin, beta 6 (209191_at), score: 0.51 TXNL4Bthioredoxin-like 4B (218794_s_at), score: -0.56 TYRP1tyrosinase-related protein 1 (205694_at), score: -0.7 U2AF1U2 small nuclear RNA auxiliary factor 1 (202858_at), score: -0.62 UAP1UDP-N-acteylglucosamine pyrophosphorylase 1 (209340_at), score: 0.54 UBA52ubiquitin A-52 residue ribosomal protein fusion product 1 (221700_s_at), score: 0.5 UBE2L6ubiquitin-conjugating enzyme E2L 6 (201649_at), score: -0.57 UBL3ubiquitin-like 3 (201535_at), score: 0.72 UFD1Lubiquitin fusion degradation 1 like (yeast) (209103_s_at), score: -0.56 UGT1A1UDP glucuronosyltransferase 1 family, polypeptide A1 (207126_x_at), score: -0.79 UGT1A6UDP glucuronosyltransferase 1 family, polypeptide A6 (206094_x_at), score: -0.73 UGT1A8UDP glucuronosyltransferase 1 family, polypeptide A8 (221304_at), score: -0.55 UGT1A9UDP glucuronosyltransferase 1 family, polypeptide A9 (204532_x_at), score: -0.75 ULK2unc-51-like kinase 2 (C. elegans) (204062_s_at), score: 0.59 URODuroporphyrinogen decarboxylase (208971_at), score: -0.61 UTYubiquitously transcribed tetratricopeptide repeat gene, Y-linked (211149_at), score: 0.53 VEGFCvascular endothelial growth factor C (209946_at), score: 0.52 VGLL3vestigial like 3 (Drosophila) (220327_at), score: 0.59 WBSCR22Williams Beuren syndrome chromosome region 22 (207628_s_at), score: -0.65 WDR74WD repeat domain 74 (221712_s_at), score: -0.65 WSB2WD repeat and SOCS box-containing 2 (201760_s_at), score: 0.5 WT1Wilms tumor 1 (206067_s_at), score: -0.92 YBX1Y box binding protein 1 (208627_s_at), score: -0.61 YPEL5yippee-like 5 (Drosophila) (217783_s_at), score: 0.58 ZFRzinc finger RNA binding protein (33148_at), score: -0.55 ZNF140zinc finger protein 140 (204523_at), score: 0.49 ZNF281zinc finger protein 281 (218401_s_at), score: 0.48 ZNF329zinc finger protein 329 (219765_at), score: 0.52 ZNF394zinc finger protein 394 (214714_at), score: -0.56
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-3407-raw-cel-1437949750.cel | 6 | 4 | Cockayne | hgu133a | CS | CSB |
| E-GEOD-3407-raw-cel-1437949579.cel | 2 | 4 | Cockayne | hgu133a | none | CSB |
| E-GEOD-3407-raw-cel-1437949721.cel | 5 | 4 | Cockayne | hgu133a | CS | CSB |
| E-GEOD-4219-raw-cel-1311956614.cel | 18 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956321.cel | 9 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956138.cel | 4 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-3407-raw-cel-1437949938.cel | 8 | 4 | Cockayne | hgu133a | none | CSB |
| E-GEOD-4219-raw-cel-1311956457.cel | 14 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956275.cel | 8 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956398.cel | 12 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956418.cel | 13 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956358.cel | 10 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956824.cel | 24 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956178.cel | 6 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |