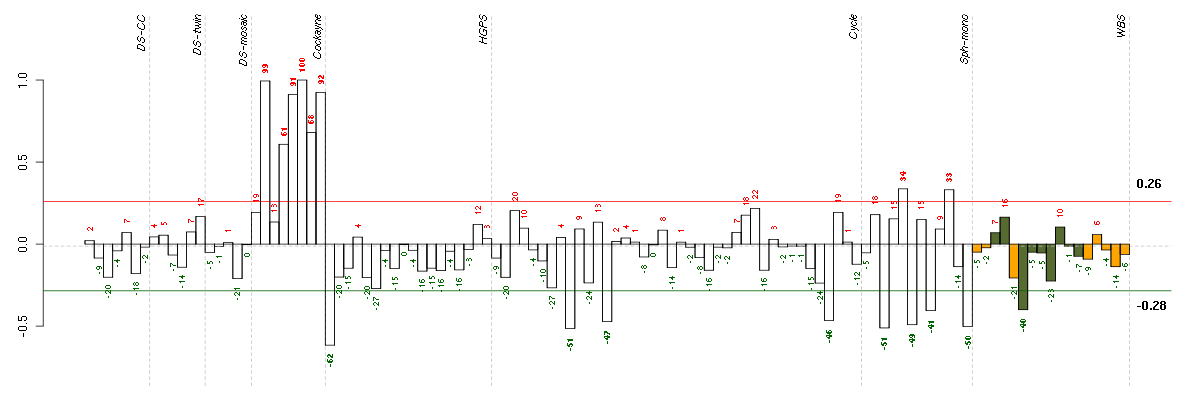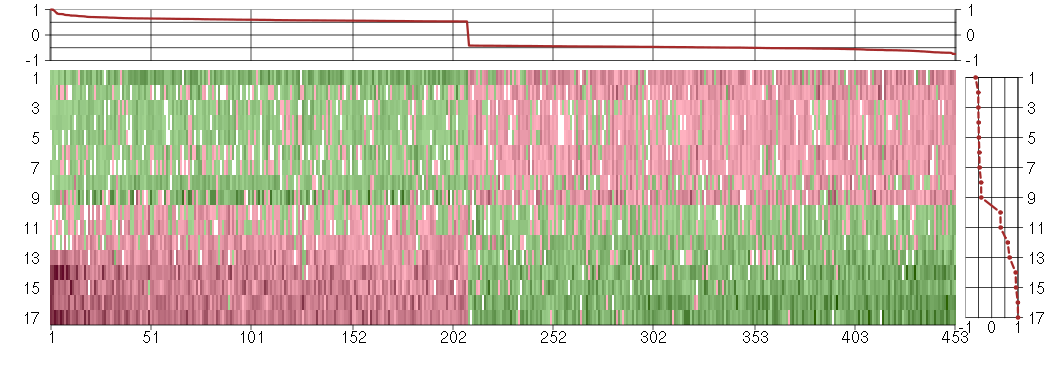

Under-expression is coded with green,
over-expression with red color.



molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
binding
The selective, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
metal ion binding
Interacting selectively with any metal ion.
cadmium ion binding
Interacting selectively with cadmium (Cd) ions.
ion binding
Interacting selectively with ions, charged atoms or groups of atoms.
cation binding
Interacting selectively with cations, charged atoms or groups of atoms with a net positive charge.
transition metal ion binding
Interacting selectively with a transition metal ions; a transition metal is an element whose atom has an incomplete d-subshell of extranuclear electrons, or which gives rise to a cation or cations with an incomplete d-subshell. Transition metals often have more than one valency state. Biologically relevant transition metals include vanadium, manganese, iron, copper, cobalt, nickel, molybdenum and silver.
all
This term is the most general term possible
transition metal ion binding
Interacting selectively with a transition metal ions; a transition metal is an element whose atom has an incomplete d-subshell of extranuclear electrons, or which gives rise to a cation or cations with an incomplete d-subshell. Transition metals often have more than one valency state. Biologically relevant transition metals include vanadium, manganese, iron, copper, cobalt, nickel, molybdenum and silver.
AASDHPPTaminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase (202170_s_at), score: 0.56 ABAT4-aminobutyrate aminotransferase (209459_s_at), score: -0.48 ABCB1ATP-binding cassette, sub-family B (MDR/TAP), member 1 (209993_at), score: 1 ABCB4ATP-binding cassette, sub-family B (MDR/TAP), member 4 (207819_s_at), score: 0.76 ABCC10ATP-binding cassette, sub-family C (CFTR/MRP), member 10 (213485_s_at), score: -0.49 ACIN1apoptotic chromatin condensation inducer 1 (201715_s_at), score: 0.64 ACP2acid phosphatase 2, lysosomal (202767_at), score: -0.46 ACTR2ARP2 actin-related protein 2 homolog (yeast) (200727_s_at), score: 0.53 ACTR8ARP8 actin-related protein 8 homolog (yeast) (218658_s_at), score: -0.51 ADAadenosine deaminase (204639_at), score: -0.45 ADO2-aminoethanethiol (cysteamine) dioxygenase (212500_at), score: -0.53 ADORA2Badenosine A2b receptor (205891_at), score: 0.62 AGPSalkylglycerone phosphate synthase (205401_at), score: 0.61 AHI1Abelson helper integration site 1 (221569_at), score: -0.45 AIM2absent in melanoma 2 (206513_at), score: 0.61 AKAP8LA kinase (PRKA) anchor protein 8-like (218064_s_at), score: 0.69 AKR1B1aldo-keto reductase family 1, member B1 (aldose reductase) (201272_at), score: 0.57 AKR1B10aldo-keto reductase family 1, member B10 (aldose reductase) (206561_s_at), score: 0.71 ANP32Aacidic (leucine-rich) nuclear phosphoprotein 32 family, member A (201043_s_at), score: 0.64 AOF2amine oxidase (flavin containing) domain 2 (212348_s_at), score: 0.63 APCadenomatous polyposis coli (203525_s_at), score: -0.47 APH1Banterior pharynx defective 1 homolog B (C. elegans) (221036_s_at), score: -0.43 APOBEC3Fapolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3F (214995_s_at), score: 0.63 APOBEC3Gapolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3G (204205_at), score: 0.63 APOL3apolipoprotein L, 3 (221087_s_at), score: 0.65 ARHGAP1Rho GTPase activating protein 1 (202117_at), score: -0.44 ARMCX3armadillo repeat containing, X-linked 3 (217858_s_at), score: -0.5 ARNTLaryl hydrocarbon receptor nuclear translocator-like (209824_s_at), score: -0.54 ARPP-19cyclic AMP phosphoprotein, 19 kD (214553_s_at), score: 0.53 ARSJarylsulfatase family, member J (219973_at), score: -0.55 ATMINATM interactor (201855_s_at), score: -0.47 ATP10AATPase, class V, type 10A (214255_at), score: -0.45 ATP11BATPase, class VI, type 11B (212536_at), score: -0.56 ATP6V1B2ATPase, H+ transporting, lysosomal 56/58kDa, V1 subunit B2 (201089_at), score: 0.58 ATP6V1FATPase, H+ transporting, lysosomal 14kDa, V1 subunit F (201527_at), score: 0.53 ATP9AATPase, class II, type 9A (212062_at), score: -0.45 BAALCbrain and acute leukemia, cytoplasmic (218899_s_at), score: -0.47 BAT1HLA-B associated transcript 1 (200041_s_at), score: 0.64 BAZ2Abromodomain adjacent to zinc finger domain, 2A (201353_s_at), score: 0.65 BCLAF1BCL2-associated transcription factor 1 (201101_s_at), score: 0.6 BHMT2betaine-homocysteine methyltransferase 2 (219902_at), score: 0.68 BMP6bone morphogenetic protein 6 (206176_at), score: -0.53 C10orf116chromosome 10 open reading frame 116 (203571_s_at), score: -0.67 C11orf17chromosome 11 open reading frame 17 (219953_s_at), score: 0.64 C11orf41chromosome 11 open reading frame 41 (214772_at), score: -0.49 C12orf47chromosome 12 open reading frame 47 (64432_at), score: 0.56 C14orf45chromosome 14 open reading frame 45 (220173_at), score: -0.6 C17orf70chromosome 17 open reading frame 70 (221800_s_at), score: -0.48 C17orf91chromosome 17 open reading frame 91 (214696_at), score: -0.48 C1orf107chromosome 1 open reading frame 107 (214193_s_at), score: -0.44 C20orf149chromosome 20 open reading frame 149 (218010_x_at), score: -0.45 C21orf66chromosome 21 open reading frame 66 (221158_at), score: 0.57 C3orf64chromosome 3 open reading frame 64 (221935_s_at), score: -0.7 C4orf30chromosome 4 open reading frame 30 (219717_at), score: 0.56 C4orf31chromosome 4 open reading frame 31 (219747_at), score: -0.52 C5orf23chromosome 5 open reading frame 23 (219054_at), score: -0.53 C5orf30chromosome 5 open reading frame 30 (221823_at), score: -0.6 CA3carbonic anhydrase III, muscle specific (204865_at), score: 0.57 CARD10caspase recruitment domain family, member 10 (210026_s_at), score: 0.56 CCNL2cyclin L2 (221427_s_at), score: 0.64 CDC25Bcell division cycle 25 homolog B (S. pombe) (201853_s_at), score: 0.58 CDC25Ccell division cycle 25 homolog C (S. pombe) (205167_s_at), score: 0.64 CDK5R1cyclin-dependent kinase 5, regulatory subunit 1 (p35) (204995_at), score: -0.46 CDKN1Acyclin-dependent kinase inhibitor 1A (p21, Cip1) (202284_s_at), score: -0.51 CDR2Lcerebellar degeneration-related protein 2-like (213230_at), score: -0.57 CHMchoroideremia (Rab escort protein 1) (207099_s_at), score: 0.53 CHPcalcium binding protein P22 (214665_s_at), score: -0.45 CKLFchemokine-like factor (219161_s_at), score: 0.55 CLINT1clathrin interactor 1 (201768_s_at), score: 0.65 COL11A1collagen, type XI, alpha 1 (37892_at), score: -0.52 COMPcartilage oligomeric matrix protein (205713_s_at), score: -0.48 COPS7ACOP9 constitutive photomorphogenic homolog subunit 7A (Arabidopsis) (209029_at), score: 0.58 COPZ2coatomer protein complex, subunit zeta 2 (219561_at), score: -0.47 CRATcarnitine acetyltransferase (209522_s_at), score: -0.45 CREBL2cAMP responsive element binding protein-like 2 (201990_s_at), score: -0.5 CRTAPcartilage associated protein (201380_at), score: -0.63 CRYABcrystallin, alpha B (209283_at), score: -0.56 CRYBG3beta-gamma crystallin domain containing 3 (214030_at), score: -0.46 CSF2RBcolony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) (205159_at), score: -0.44 CTBP1C-terminal binding protein 1 (213979_s_at), score: 0.56 CTNNBL1catenin, beta like 1 (221021_s_at), score: 0.64 CTTNcortactin (201059_at), score: -0.42 CUL3cullin 3 (201370_s_at), score: -0.44 CUX1cut-like homeobox 1 (214743_at), score: -0.45 CYBAcytochrome b-245, alpha polypeptide (203028_s_at), score: -0.42 DBF4DBF4 homolog (S. cerevisiae) (204244_s_at), score: 0.53 DDAH1dimethylarginine dimethylaminohydrolase 1 (209094_at), score: -0.52 DDX17DEAD (Asp-Glu-Ala-Asp) box polypeptide 17 (208151_x_at), score: 0.79 DDX3XDEAD (Asp-Glu-Ala-Asp) box polypeptide 3, X-linked (201211_s_at), score: 0.63 DDX3YDEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked (205000_at), score: -0.61 DGKDdiacylglycerol kinase, delta 130kDa (208072_s_at), score: 0.65 DHX35DEAH (Asp-Glu-Ala-His) box polypeptide 35 (218579_s_at), score: 0.53 DHX38DEAH (Asp-Glu-Ala-His) box polypeptide 38 (209178_at), score: 0.66 DHX9DEAH (Asp-Glu-Ala-His) box polypeptide 9 (212105_s_at), score: 0.68 DIO2deiodinase, iodothyronine, type II (203699_s_at), score: 0.78 DMWDdystrophia myotonica, WD repeat containing (213231_at), score: 0.6 DNAJB1DnaJ (Hsp40) homolog, subfamily B, member 1 (200664_s_at), score: 0.57 DNAJC7DnaJ (Hsp40) homolog, subfamily C, member 7 (202416_at), score: 0.55 DRAP1DR1-associated protein 1 (negative cofactor 2 alpha) (203258_at), score: 0.62 DSPdesmoplakin (200606_at), score: -0.49 DUSP14dual specificity phosphatase 14 (203367_at), score: -0.62 ECSITECSIT homolog (Drosophila) (218225_at), score: 0.57 EDEM1ER degradation enhancer, mannosidase alpha-like 1 (203279_at), score: -0.45 EDNRAendothelin receptor type A (204464_s_at), score: -0.55 EIF1AYeukaryotic translation initiation factor 1A, Y-linked (204409_s_at), score: -0.51 EMP1epithelial membrane protein 1 (201325_s_at), score: -0.42 EPB41L2erythrocyte membrane protein band 4.1-like 2 (201718_s_at), score: 0.55 EPHB2EPH receptor B2 (209589_s_at), score: 0.69 EPHX1epoxide hydrolase 1, microsomal (xenobiotic) (202017_at), score: 0.55 ERO1LERO1-like (S. cerevisiae) (218498_s_at), score: -0.52 ESM1endothelial cell-specific molecule 1 (208394_x_at), score: -0.59 ETS1v-ets erythroblastosis virus E26 oncogene homolog 1 (avian) (214447_at), score: 0.61 EWSR1Ewing sarcoma breakpoint region 1 (209214_s_at), score: 0.63 FAF2Fas associated factor family member 2 (212106_at), score: 0.57 FAM105Afamily with sequence similarity 105, member A (219694_at), score: 0.61 FAM108A1family with sequence similarity 108, member A1 (221267_s_at), score: -0.47 FAM155Afamily with sequence similarity 155, member A (214825_at), score: -0.45 FAM168Bfamily with sequence similarity 168, member B (212017_at), score: 0.55 FAM169Afamily with sequence similarity 169, member A (213954_at), score: 0.67 FAM63Bfamily with sequence similarity 63, member B (214691_x_at), score: -0.45 FAM8A1family with sequence similarity 8, member A1 (203420_at), score: -0.58 FAT1FAT tumor suppressor homolog 1 (Drosophila) (201579_at), score: -0.7 FBN2fibrillin 2 (203184_at), score: -0.64 FKBP9FK506 binding protein 9, 63 kDa (212169_at), score: -0.59 FLJ11151hypothetical protein FLJ11151 (218610_s_at), score: -0.46 FLJ12529pre-mRNA cleavage factor I, 59 kDa subunit (217866_at), score: 0.63 FLRT2fibronectin leucine rich transmembrane protein 2 (204359_at), score: -0.48 FNDC3Afibronectin type III domain containing 3A (202304_at), score: -0.46 FOXF2forkhead box F2 (206377_at), score: 0.54 FOXG1forkhead box G1 (206018_at), score: 0.59 FOXL2forkhead box L2 (220102_at), score: 0.57 FUSfusion (involved in t(12;16) in malignant liposarcoma) (217370_x_at), score: 0.61 FXR1fragile X mental retardation, autosomal homolog 1 (201635_s_at), score: 0.59 GABBR2gamma-aminobutyric acid (GABA) B receptor, 2 (209990_s_at), score: -0.47 GABREgamma-aminobutyric acid (GABA) A receptor, epsilon (204537_s_at), score: 0.6 GAGE4G antigen 4 (207086_x_at), score: 0.93 GAGE6G antigen 6 (208155_x_at), score: 0.82 GATA3GATA binding protein 3 (209604_s_at), score: 0.75 GBP1guanylate binding protein 1, interferon-inducible, 67kDa (202269_x_at), score: 0.59 GHRgrowth hormone receptor (205498_at), score: -0.66 GLE1GLE1 RNA export mediator homolog (yeast) (206920_s_at), score: 0.68 GLSglutaminase (203159_at), score: -0.6 GLT25D2glycosyltransferase 25 domain containing 2 (209883_at), score: -0.52 GLTPglycolipid transfer protein (219267_at), score: -0.57 GMDSGDP-mannose 4,6-dehydratase (204875_s_at), score: 0.57 GMPRguanosine monophosphate reductase (204187_at), score: 0.54 GNA12guanine nucleotide binding protein (G protein) alpha 12 (221737_at), score: -0.52 GPR137BG protein-coupled receptor 137B (204137_at), score: -0.49 GRAMD3GRAM domain containing 3 (218706_s_at), score: -0.43 GREM1gremlin 1, cysteine knot superfamily, homolog (Xenopus laevis) (218469_at), score: -0.42 GSPT2G1 to S phase transition 2 (205541_s_at), score: -0.5 GTF2H4general transcription factor IIH, polypeptide 4, 52kDa (203577_at), score: 0.53 H2AFYH2A histone family, member Y (214500_at), score: 0.53 HAS2hyaluronan synthase 2 (206432_at), score: -0.54 HEPHhephaestin (203903_s_at), score: -0.55 HERC5hect domain and RLD 5 (219863_at), score: 0.82 HGSNATheparan-alpha-glucosaminide N-acetyltransferase (218017_s_at), score: -0.68 HIPK1homeodomain interacting protein kinase 1 (212291_at), score: 0.57 HIST1H4Chistone cluster 1, H4c (205967_at), score: -0.46 HIVEP2human immunodeficiency virus type I enhancer binding protein 2 (212642_s_at), score: -0.54 HJURPHolliday junction recognition protein (218726_at), score: 0.54 HLA-Bmajor histocompatibility complex, class I, B (211911_x_at), score: 0.6 HMGB1L10high-mobility group box 1-like 10 (216508_x_at), score: 0.6 HNRNPCheterogeneous nuclear ribonucleoprotein C (C1/C2) (200751_s_at), score: 0.6 HNRNPH1heterogeneous nuclear ribonucleoprotein H1 (H) (213470_s_at), score: 0.58 HNRNPUheterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A) (200593_s_at), score: 0.58 HOXA10homeobox A10 (213150_at), score: -0.46 HRASLSHRAS-like suppressor (219983_at), score: 0.56 HSD17B12hydroxysteroid (17-beta) dehydrogenase 12 (217869_at), score: -0.52 HSP90AB1heat shock protein 90kDa alpha (cytosolic), class B member 1 (214359_s_at), score: 0.62 HSPA4heat shock 70kDa protein 4 (211016_x_at), score: 0.59 HSPB2heat shock 27kDa protein 2 (205824_at), score: -0.42 HSPB7heat shock 27kDa protein family, member 7 (cardiovascular) (218934_s_at), score: -0.6 IDEinsulin-degrading enzyme (203327_at), score: -0.42 IDH3Bisocitrate dehydrogenase 3 (NAD+) beta (210014_x_at), score: 0.54 IFI16interferon, gamma-inducible protein 16 (208965_s_at), score: 0.58 IFIT3interferon-induced protein with tetratricopeptide repeats 3 (204747_at), score: 0.66 IGF2BP3insulin-like growth factor 2 mRNA binding protein 3 (203819_s_at), score: 0.56 IGFBP3insulin-like growth factor binding protein 3 (212143_s_at), score: -0.47 IGSF3immunoglobulin superfamily, member 3 (202421_at), score: 0.61 IKIK cytokine, down-regulator of HLA II (200066_at), score: 0.64 IL11interleukin 11 (206924_at), score: -0.44 IL21Rinterleukin 21 receptor (219971_at), score: -0.43 ILF3interleukin enhancer binding factor 3, 90kDa (208930_s_at), score: 0.55 IPO8importin 8 (205701_at), score: 0.54 IRS2insulin receptor substrate 2 (209185_s_at), score: -0.53 ISLRimmunoglobulin superfamily containing leucine-rich repeat (207191_s_at), score: -0.46 JAK2Janus kinase 2 (a protein tyrosine kinase) (205842_s_at), score: 0.55 JARID1Djumonji, AT rich interactive domain 1D (206700_s_at), score: -0.54 KCTD12potassium channel tetramerisation domain containing 12 (212192_at), score: -0.44 KIAA0355KIAA0355 (203288_at), score: -0.48 KIAA0495KIAA0495 (213340_s_at), score: -0.55 KIF2Ckinesin family member 2C (209408_at), score: 0.53 KIF3Akinesin family member 3A (213623_at), score: 0.6 KIF3Bkinesin family member 3B (203943_at), score: 0.53 KIFC1kinesin family member C1 (209680_s_at), score: 0.66 KLF4Kruppel-like factor 4 (gut) (221841_s_at), score: -0.43 KRIT1KRIT1, ankyrin repeat containing (34031_i_at), score: 0.55 KRT19keratin 19 (201650_at), score: -0.44 KYNUkynureninase (L-kynurenine hydrolase) (217388_s_at), score: 0.58 LAMB3laminin, beta 3 (209270_at), score: 0.73 LAS1LLAS1-like (S. cerevisiae) (208117_s_at), score: 0.55 LEF1lymphoid enhancer-binding factor 1 (221558_s_at), score: 0.68 LGMNlegumain (201212_at), score: -0.51 LIMK2LIM domain kinase 2 (202193_at), score: -0.46 LIPAlipase A, lysosomal acid, cholesterol esterase (201847_at), score: -0.58 LMO7LIM domain 7 (202674_s_at), score: -0.42 LOC151162hypothetical LOC151162 (212098_at), score: -0.49 LOC388796hypothetical LOC388796 (65588_at), score: 0.59 LOC728855hypothetical LOC728855 (222001_x_at), score: 0.7 LOC730092RRN3 RNA polymerase I transcription factor homolog (S. cerevisiae) pseudogene (216908_x_at), score: 0.53 LOC90379hypothetical protein BC002926 (221849_s_at), score: 0.56 LRIG2leucine-rich repeats and immunoglobulin-like domains 2 (205953_at), score: 0.57 LRRC2leucine rich repeat containing 2 (219949_at), score: -0.63 LRRN3leucine rich repeat neuronal 3 (209841_s_at), score: -0.43 LTBP2latent transforming growth factor beta binding protein 2 (204682_at), score: -0.61 LUMlumican (201744_s_at), score: -0.54 M6PRmannose-6-phosphate receptor (cation dependent) (200900_s_at), score: 0.66 MAGI1membrane associated guanylate kinase, WW and PDZ domain containing 1 (206144_at), score: -0.43 MAGOHmago-nashi homolog, proliferation-associated (Drosophila) (210092_at), score: 0.62 MALLmal, T-cell differentiation protein-like (209373_at), score: -0.44 MAN2A1mannosidase, alpha, class 2A, member 1 (205105_at), score: -0.64 MARCH2membrane-associated ring finger (C3HC4) 2 (210075_at), score: -0.67 MARK1MAP/microtubule affinity-regulating kinase 1 (221047_s_at), score: 0.67 MBOAT2membrane bound O-acyltransferase domain containing 2 (213288_at), score: -0.49 MED6mediator complex subunit 6 (207079_s_at), score: 0.58 MFAP3Lmicrofibrillar-associated protein 3-like (205442_at), score: -0.56 MFAP5microfibrillar associated protein 5 (213764_s_at), score: -0.6 MGEA5meningioma expressed antigen 5 (hyaluronidase) (200898_s_at), score: 0.56 MMP1matrix metallopeptidase 1 (interstitial collagenase) (204475_at), score: -0.48 MMP16matrix metallopeptidase 16 (membrane-inserted) (207012_at), score: -0.47 MOXD1monooxygenase, DBH-like 1 (209708_at), score: -0.42 MRC2mannose receptor, C type 2 (37408_at), score: -0.43 MREGmelanoregulin (219648_at), score: -0.49 MRPL28mitochondrial ribosomal protein L28 (204599_s_at), score: 0.53 MRPL9mitochondrial ribosomal protein L9 (211594_s_at), score: 0.58 MT1Fmetallothionein 1F (217165_x_at), score: -0.44 MT1Gmetallothionein 1G (204745_x_at), score: -0.49 MT1Hmetallothionein 1H (206461_x_at), score: -0.42 MT1Mmetallothionein 1M (217546_at), score: -0.58 MT1P3metallothionein 1 pseudogene 3 (221953_s_at), score: -0.43 MT1Xmetallothionein 1X (204326_x_at), score: -0.45 MXRA5matrix-remodelling associated 5 (209596_at), score: -0.46 MXRA8matrix-remodelling associated 8 (213422_s_at), score: -0.48 MYO10myosin X (201976_s_at), score: -0.55 MYO1Cmyosin IC (214656_x_at), score: -0.69 MYO1Dmyosin ID (212338_at), score: -0.49 NACC2NACC family member 2, BEN and BTB (POZ) domain containing (212993_at), score: -0.47 NAPGN-ethylmaleimide-sensitive factor attachment protein, gamma (210048_at), score: 0.6 NASPnuclear autoantigenic sperm protein (histone-binding) (201970_s_at), score: 0.61 NBASneuroblastoma amplified sequence (202926_at), score: -0.42 NCALDneurocalcin delta (211685_s_at), score: -0.59 NCK2NCK adaptor protein 2 (203315_at), score: 0.58 NCR2natural cytotoxicity triggering receptor 2 (217045_x_at), score: -0.42 NCRNA00084non-protein coding RNA 84 (214657_s_at), score: 0.64 NDNnecdin homolog (mouse) (209550_at), score: -0.56 NDUFS3NADH dehydrogenase (ubiquinone) Fe-S protein 3, 30kDa (NADH-coenzyme Q reductase) (201740_at), score: 0.54 NEK7NIMA (never in mitosis gene a)-related kinase 7 (212530_at), score: -0.59 NFASCneurofascin homolog (chicken) (213438_at), score: -0.42 NGDNneuroguidin, EIF4E binding protein (213794_s_at), score: 0.63 NMUneuromedin U (206023_at), score: 0.61 NOC2Lnucleolar complex associated 2 homolog (S. cerevisiae) (202115_s_at), score: 0.56 NOL12nucleolar protein 12 (219324_at), score: 0.62 NONOnon-POU domain containing, octamer-binding (208698_s_at), score: 0.74 NOTCH2Notch homolog 2 (Drosophila) (210756_s_at), score: 0.56 NOX4NADPH oxidase 4 (219773_at), score: -0.46 NPR3natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) (219789_at), score: -0.44 NRGNneurogranin (protein kinase C substrate, RC3) (204081_at), score: 0.66 NUP93nucleoporin 93kDa (202188_at), score: 0.54 OAS32'-5'-oligoadenylate synthetase 3, 100kDa (218400_at), score: 0.62 OASL2'-5'-oligoadenylate synthetase-like (210797_s_at), score: 0.99 ODF2outer dense fiber of sperm tails 2 (210415_s_at), score: 0.53 ODZ4odz, odd Oz/ten-m homolog 4 (Drosophila) (213273_at), score: -0.47 OPCMLopioid binding protein/cell adhesion molecule-like (214111_at), score: -0.6 OXA1Loxidase (cytochrome c) assembly 1-like (208717_at), score: 0.58 OXTRoxytocin receptor (206825_at), score: -0.45 P2RX5purinergic receptor P2X, ligand-gated ion channel, 5 (210448_s_at), score: -0.42 P4HA1prolyl 4-hydroxylase, alpha polypeptide I (207543_s_at), score: -0.51 PARP4poly (ADP-ribose) polymerase family, member 4 (202239_at), score: 0.56 PBX1pre-B-cell leukemia homeobox 1 (212148_at), score: -0.49 PCDH9protocadherin 9 (219737_s_at), score: -0.48 PCLOpiccolo (presynaptic cytomatrix protein) (213558_at), score: 0.65 PCTK2PCTAIRE protein kinase 2 (221918_at), score: -0.61 PDAP1PDGFA associated protein 1 (202290_at), score: 0.63 PDCD1LG2programmed cell death 1 ligand 2 (220049_s_at), score: -0.43 PDGFCplatelet derived growth factor C (218718_at), score: -0.43 PDPK13-phosphoinositide dependent protein kinase-1 (204524_at), score: -0.54 PER3period homolog 3 (Drosophila) (221045_s_at), score: 0.54 PGRMC2progesterone receptor membrane component 2 (213227_at), score: -0.49 PID1phosphotyrosine interaction domain containing 1 (219093_at), score: -0.54 PILRBpaired immunoglobin-like type 2 receptor beta (220954_s_at), score: 0.76 PIWIL1piwi-like 1 (Drosophila) (214868_at), score: 0.55 PLK2polo-like kinase 2 (Drosophila) (201939_at), score: 0.64 POM121POM121 membrane glycoprotein (rat) (212178_s_at), score: 0.56 PPIHpeptidylprolyl isomerase H (cyclophilin H) (204228_at), score: 0.65 PPM2Cprotein phosphatase 2C, magnesium-dependent, catalytic subunit (218273_s_at), score: 0.6 PPP1R3Cprotein phosphatase 1, regulatory (inhibitor) subunit 3C (204284_at), score: -0.48 PPP1R3Dprotein phosphatase 1, regulatory (inhibitor) subunit 3D (204554_at), score: -0.56 PPP3CBprotein phosphatase 3 (formerly 2B), catalytic subunit, beta isoform (209817_at), score: -0.54 PQBP1polyglutamine binding protein 1 (214527_s_at), score: 0.67 PRPF4PRP4 pre-mRNA processing factor 4 homolog (yeast) (209162_s_at), score: 0.59 PSMB8proteasome (prosome, macropain) subunit, beta type, 8 (large multifunctional peptidase 7) (209040_s_at), score: 0.74 PSMC3proteasome (prosome, macropain) 26S subunit, ATPase, 3 (201267_s_at), score: 0.61 PSMD4proteasome (prosome, macropain) 26S subunit, non-ATPase, 4 (211609_x_at), score: 0.55 PTGS2prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) (204748_at), score: -0.52 PTHLHparathyroid hormone-like hormone (211756_at), score: -0.59 PTPRBprotein tyrosine phosphatase, receptor type, B (205846_at), score: -0.49 PXDNperoxidasin homolog (Drosophila) (212013_at), score: -0.52 QPCTglutaminyl-peptide cyclotransferase (205174_s_at), score: -0.53 R3HDM2R3H domain containing 2 (203831_at), score: -0.53 RAB1ARAB1A, member RAS oncogene family (213440_at), score: -0.52 RAB6CRAB6C, member RAS oncogene family (210406_s_at), score: -0.51 RAB7L1RAB7, member RAS oncogene family-like 1 (218700_s_at), score: 0.7 RABGAP1LRAB GTPase activating protein 1-like (213982_s_at), score: -0.49 RB1retinoblastoma 1 (203132_at), score: -0.45 RBM12RNA binding motif protein 12 (212168_at), score: 0.57 RCBTB2regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 (204759_at), score: -0.47 RECKreversion-inducing-cysteine-rich protein with kazal motifs (205407_at), score: -0.52 RGP1RGP1 retrograde golgi transport homolog (S. cerevisiae) (203169_at), score: 0.54 RHODras homolog gene family, member D (31846_at), score: -0.48 RIN2Ras and Rab interactor 2 (209684_at), score: -0.49 RNF19Aring finger protein 19A (220483_s_at), score: 0.61 RNF220ring finger protein 220 (219988_s_at), score: 0.56 RNPS1RNA binding protein S1, serine-rich domain (207939_x_at), score: 0.54 RPL31ribosomal protein L31 (221593_s_at), score: -0.52 RPP14ribonuclease P/MRP 14kDa subunit (204245_s_at), score: 0.64 RPS20ribosomal protein S20 (216247_at), score: -0.42 RPS4Y1ribosomal protein S4, Y-linked 1 (201909_at), score: -0.43 S1PR1sphingosine-1-phosphate receptor 1 (204642_at), score: -0.46 SAMD4Asterile alpha motif domain containing 4A (212845_at), score: -0.52 SART3squamous cell carcinoma antigen recognized by T cells 3 (209127_s_at), score: 0.69 SC65synaptonemal complex protein SC65 (204078_at), score: -0.5 SCG5secretogranin V (7B2 protein) (203889_at), score: -0.45 SEC16ASEC16 homolog A (S. cerevisiae) (215696_s_at), score: -0.46 SEMA3Fsema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F (35666_at), score: -0.41 SERPINB7serpin peptidase inhibitor, clade B (ovalbumin), member 7 (206421_s_at), score: -0.44 SERPINE2serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 (212190_at), score: -0.45 SF3B1splicing factor 3b, subunit 1, 155kDa (201070_x_at), score: 0.53 SF4splicing factor 4 (215004_s_at), score: 0.56 SFPQsplicing factor proline/glutamine-rich (polypyrimidine tract binding protein associated) (201585_s_at), score: 0.58 SFXN3sideroflexin 3 (220974_x_at), score: -0.49 SGCDsarcoglycan, delta (35kDa dystrophin-associated glycoprotein) (213543_at), score: -0.52 SH3YL1SH3 domain containing, Ysc84-like 1 (S. cerevisiae) (204019_s_at), score: -0.46 SHANK2SH3 and multiple ankyrin repeat domains 2 (213307_at), score: -0.43 SHFM1split hand/foot malformation (ectrodactyly) type 1 (202276_at), score: 0.58 SHMT1serine hydroxymethyltransferase 1 (soluble) (209980_s_at), score: 0.55 SIDT2SID1 transmembrane family, member 2 (56256_at), score: -0.5 SLC16A1solute carrier family 16, member 1 (monocarboxylic acid transporter 1) (209900_s_at), score: -0.65 SLC1A1solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1 (213664_at), score: -0.47 SLC25A32solute carrier family 25, member 32 (221020_s_at), score: -0.53 SLC25A37solute carrier family 25, member 37 (218136_s_at), score: 0.59 SLC2A10solute carrier family 2 (facilitated glucose transporter), member 10 (221024_s_at), score: -0.54 SLC30A1solute carrier family 30 (zinc transporter), member 1 (212907_at), score: -0.74 SLC35A3solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3 (206770_s_at), score: -0.53 SLC39A4solute carrier family 39 (zinc transporter), member 4 (219215_s_at), score: -0.44 SLC4A4solute carrier family 4, sodium bicarbonate cotransporter, member 4 (203908_at), score: -0.68 SLC7A1solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 (212295_s_at), score: -0.44 SLC7A11solute carrier family 7, (cationic amino acid transporter, y+ system) member 11 (217678_at), score: 0.65 SMAD3SMAD family member 3 (218284_at), score: 0.53 SMAD5SMAD family member 5 (205187_at), score: 0.61 SMC3structural maintenance of chromosomes 3 (209257_s_at), score: 0.55 SMEK1SMEK homolog 1, suppressor of mek1 (Dictyostelium) (220368_s_at), score: 0.7 SMG1SMG1 homolog, phosphatidylinositol 3-kinase-related kinase (C. elegans) (210057_at), score: 0.71 SMURF2SMAD specific E3 ubiquitin protein ligase 2 (205596_s_at), score: -0.44 SNCAsynuclein, alpha (non A4 component of amyloid precursor) (204466_s_at), score: 0.76 SNRNP27small nuclear ribonucleoprotein 27kDa (U4/U6.U5) (212440_at), score: -0.46 SNW1SNW domain containing 1 (215424_s_at), score: 0.54 SOS2son of sevenless homolog 2 (Drosophila) (212870_at), score: -0.47 SP110SP110 nuclear body protein (209761_s_at), score: 0.55 SPARCsecreted protein, acidic, cysteine-rich (osteonectin) (212667_at), score: -0.45 SPHARS-phase response (cyclin-related) (206272_at), score: -0.58 SPINT2serine peptidase inhibitor, Kunitz type, 2 (210715_s_at), score: -0.49 SQSTM1sequestosome 1 (201471_s_at), score: 0.58 SS18synovial sarcoma translocation, chromosome 18 (216684_s_at), score: 0.57 SSNA1Sjogren syndrome nuclear autoantigen 1 (210378_s_at), score: 0.56 SSR3signal sequence receptor, gamma (translocon-associated protein gamma) (217790_s_at), score: -0.7 SSRP1structure specific recognition protein 1 (200956_s_at), score: 0.7 STOML2stomatin (EPB72)-like 2 (215416_s_at), score: 0.61 STSsteroid sulfatase (microsomal), isozyme S (203767_s_at), score: -0.43 STX12syntaxin 12 (212112_s_at), score: -0.52 STX16syntaxin 16 (221499_s_at), score: 0.57 STX2syntaxin 2 (213434_at), score: -0.45 SULF1sulfatase 1 (212353_at), score: -0.51 SUPT16Hsuppressor of Ty 16 homolog (S. cerevisiae) (217815_at), score: 0.57 SYTL2synaptotagmin-like 2 (220613_s_at), score: 0.6 TACSTD2tumor-associated calcium signal transducer 2 (202286_s_at), score: -0.52 TBC1D3TBC1 domain family, member 3 (209403_at), score: 0.63 TBC1D8TBC1 domain family, member 8 (with GRAM domain) (204526_s_at), score: 0.58 TBC1D8BTBC1 domain family, member 8B (with GRAM domain) (219771_at), score: -0.48 TBX5T-box 5 (207155_at), score: -0.42 TCEAL2transcription elongation factor A (SII)-like 2 (211276_at), score: 0.73 TEKTEK tyrosine kinase, endothelial (206702_at), score: -0.52 TERF1telomeric repeat binding factor (NIMA-interacting) 1 (203448_s_at), score: 0.57 TESK1testis-specific kinase 1 (204106_at), score: -0.46 TGFAtransforming growth factor, alpha (205016_at), score: -0.45 TGIF1TGFB-induced factor homeobox 1 (203313_s_at), score: 0.53 THAP1THAP domain containing, apoptosis associated protein 1 (219292_at), score: -0.43 THSD7Athrombospondin, type I, domain containing 7A (214920_at), score: 0.54 TIMP2TIMP metallopeptidase inhibitor 2 (203167_at), score: -0.51 TKTtransketolase (208699_x_at), score: 0.6 TM9SF3transmembrane 9 superfamily member 3 (217758_s_at), score: -0.49 TMEFF1transmembrane protein with EGF-like and two follistatin-like domains 1 (205122_at), score: -0.6 TMEM2transmembrane protein 2 (218113_at), score: -0.46 TMEM204transmembrane protein 204 (219315_s_at), score: -0.61 TMEM22transmembrane protein 22 (219569_s_at), score: -0.42 TMEM47transmembrane protein 47 (209656_s_at), score: -0.58 TMEM51transmembrane protein 51 (218815_s_at), score: 0.65 TMEM97transmembrane protein 97 (212279_at), score: 0.63 TNFRSF1Atumor necrosis factor receptor superfamily, member 1A (207643_s_at), score: 0.54 TNS1tensin 1 (221748_s_at), score: -0.74 TOB1transducer of ERBB2, 1 (202704_at), score: -0.69 TRIM16tripartite motif-containing 16 (204341_at), score: 0.7 TRIM8tripartite motif-containing 8 (221012_s_at), score: -0.43 TRPC4transient receptor potential cation channel, subfamily C, member 4 (220817_at), score: -0.43 TSNtranslin (201504_s_at), score: 0.63 TSNAXtranslin-associated factor X (203983_at), score: -0.47 TSPAN13tetraspanin 13 (217979_at), score: -0.69 TSPAN31tetraspanin 31 (203227_s_at), score: -0.47 TSPYL1TSPY-like 1 (221493_at), score: -0.63 TSPYL5TSPY-like 5 (213122_at), score: -0.68 TTTY15testis-specific transcript, Y-linked 15 (214983_at), score: -0.5 TYRP1tyrosinase-related protein 1 (205694_at), score: 0.63 U2AF1U2 small nuclear RNA auxiliary factor 1 (202858_at), score: 0.55 UAP1UDP-N-acteylglucosamine pyrophosphorylase 1 (209340_at), score: -0.62 UBE2D4ubiquitin-conjugating enzyme E2D 4 (putative) (218837_s_at), score: -0.47 UBE2Hubiquitin-conjugating enzyme E2H (UBC8 homolog, yeast) (221962_s_at), score: 0.66 UBE2L6ubiquitin-conjugating enzyme E2L 6 (201649_at), score: 0.65 UBL3ubiquitin-like 3 (201535_at), score: -0.66 UCHL1ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) (201387_s_at), score: -0.51 UGCGUDP-glucose ceramide glucosyltransferase (204881_s_at), score: 0.54 UGT1A1UDP glucuronosyltransferase 1 family, polypeptide A1 (207126_x_at), score: 0.84 UGT1A6UDP glucuronosyltransferase 1 family, polypeptide A6 (206094_x_at), score: 0.82 UGT1A9UDP glucuronosyltransferase 1 family, polypeptide A9 (204532_x_at), score: 0.78 URODuroporphyrinogen decarboxylase (208971_at), score: 0.6 USP18ubiquitin specific peptidase 18 (219211_at), score: 0.57 USP9Yubiquitin specific peptidase 9, Y-linked (fat facets-like, Drosophila) (206624_at), score: -0.5 UTP14CUTP14, U3 small nucleolar ribonucleoprotein, homolog C (yeast) (203614_at), score: -0.55 UTYubiquitously transcribed tetratricopeptide repeat gene, Y-linked (211149_at), score: -0.45 VEGFCvascular endothelial growth factor C (209946_at), score: -0.48 VLDLRvery low density lipoprotein receptor (209822_s_at), score: -0.45 VWA5Avon Willebrand factor A domain containing 5A (210102_at), score: 0.56 WDR48WD repeat domain 48 (222157_s_at), score: 0.59 WDR74WD repeat domain 74 (221712_s_at), score: 0.56 WNK1WNK lysine deficient protein kinase 1 (211993_at), score: 0.57 WT1Wilms tumor 1 (206067_s_at), score: 0.69 WWC3WWC family member 3 (219520_s_at), score: -0.49 XYLT1xylosyltransferase I (213725_x_at), score: -0.48 ZNF140zinc finger protein 140 (204523_at), score: -0.42 ZNF224zinc finger protein 224 (216983_s_at), score: 0.56 ZNF227zinc finger protein 227 (217403_s_at), score: 0.58 ZNF281zinc finger protein 281 (218401_s_at), score: -0.6 ZNF329zinc finger protein 329 (219765_at), score: -0.43 ZNF415zinc finger protein 415 (205514_at), score: -0.45
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-3860-raw-cel-1561690199.cel | 1 | 5 | HGPS | hgu133a | none | GM0316B |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-4219-raw-cel-1311956178.cel | 6 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956824.cel | 24 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956358.cel | 10 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-4219-raw-cel-1311956418.cel | 13 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| 5042_CNTL.CEL | 6 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| E-GEOD-4219-raw-cel-1311956614.cel | 18 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956321.cel | 9 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-3407-raw-cel-1437949704.cel | 4 | 4 | Cockayne | hgu133a | CS | eGFP |
| E-GEOD-3407-raw-cel-1437949854.cel | 7 | 4 | Cockayne | hgu133a | CS | eGFP |
| E-GEOD-3407-raw-cel-1437949721.cel | 5 | 4 | Cockayne | hgu133a | CS | CSB |
| E-GEOD-3407-raw-cel-1437949938.cel | 8 | 4 | Cockayne | hgu133a | none | CSB |
| E-GEOD-3407-raw-cel-1437949579.cel | 2 | 4 | Cockayne | hgu133a | none | CSB |
| E-GEOD-3407-raw-cel-1437949750.cel | 6 | 4 | Cockayne | hgu133a | CS | CSB |