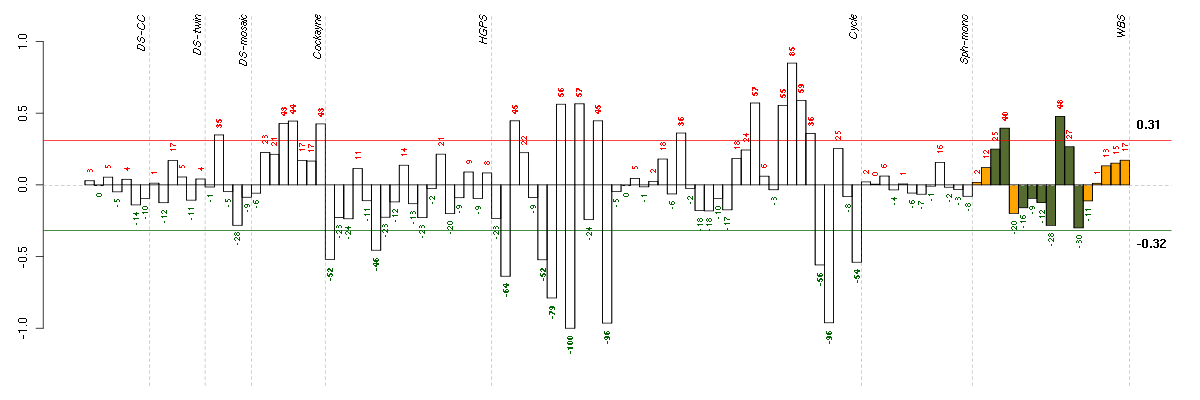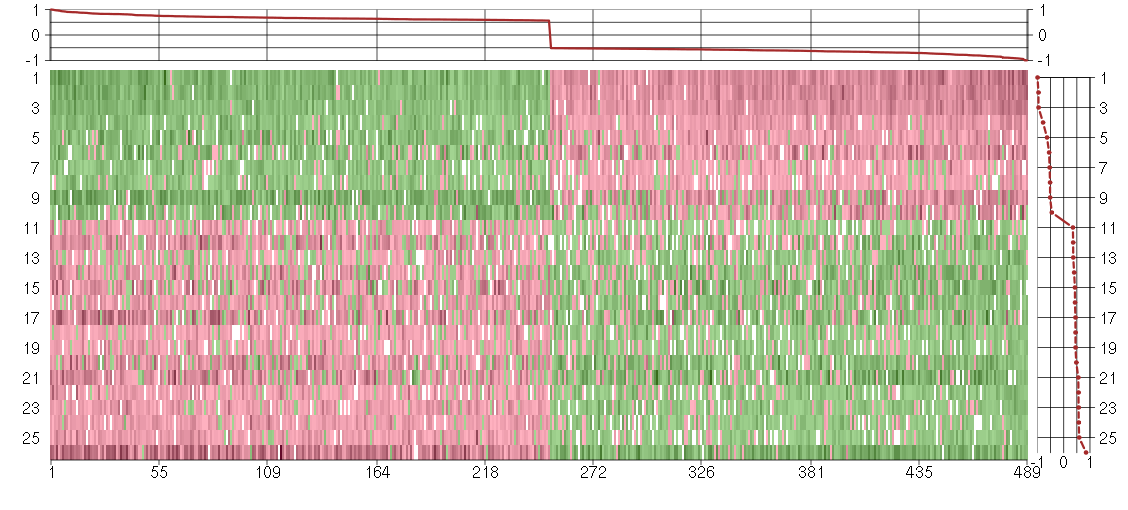

Under-expression is coded with green,
over-expression with red color.

immune system process
Any process involved in the development or functioning of the immune system, an organismal system for calibrated responses to potential internal or invasive threats.
immune system development
The process whose specific outcome is the progression of an organismal system whose objective is to provide calibrated responses by an organism to a potential internal or invasive threat, over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
leukocyte differentiation
The process whereby a relatively unspecialized hemopoietic precursor cell acquires the specialized features of a plasmacytoid dendritic cell or any cell of the myeloid leukocyte or lymphocyte lineages.
myeloid leukocyte differentiation
The process whereby a relatively unspecialized myeloid precursor cell acquires the specialized features of any cell of the myeloid leukocyte lineage.
regulation of immune system process
Any process that modulates the frequency, rate, or extent of an immune system process.
regulation of myeloid leukocyte differentiation
Any process that modulates the frequency, rate, or extent of myeloid leukocyte differentiation.
negative regulation of myeloid leukocyte differentiation
Any process that stops, prevents, or reduces the frequency, rate, or extent of myeloid leukocyte differentiation.
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cell proliferation
The multiplication or reproduction of cells, resulting in the expansion of a cell population.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
hemopoiesis
The process whose specific outcome is the progression of the myeloid and lymphoid derived organ/tissue systems of the blood and other parts of the body over time, from formation to the mature structure. The site of hemopoiesis is variable during development, but occurs primarily in bone marrow or kidney in many adult vertebrates.
myeloid cell differentiation
The process whereby a relatively unspecialized myeloid precursor cell acquires the specialized features of any cell of the myeloid leukocyte, megakaryocyte, thrombocyte, or erythrocyte lineages.
cell differentiation
The process whereby relatively unspecialized cells, e.g. embryonic or regenerative cells, acquire specialized structural and/or functional features that characterize the cells, tissues, or organs of the mature organism or some other relatively stable phase of the organism's life history. Differentiation includes the processes involved in commitment of a cell to a specific fate.
macrophage differentiation
The process whereby a relatively unspecialized monocyte acquires the specialized features of a macrophage.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
developmental process
A biological process whose specific outcome is the progression of an integrated living unit: an anatomical structure (which may be a subcellular structure, cell, tissue, or organ), or organism over time from an initial condition to a later condition.
regulation of cell proliferation
Any process that modulates the frequency, rate or extent of cell proliferation.
regulation of cell differentiation
Any process that modulates the frequency, rate or extent of cell differentiation, the process whereby relatively unspecialized cells acquire specialized structural and functional features.
negative regulation of cell differentiation
Any process that stops, prevents or reduces the frequency, rate or extent of cell differentiation.
regulation of myeloid cell differentiation
Any process that modulates the frequency, rate or extent of myeloid cell differentiation.
negative regulation of myeloid cell differentiation
Any process that stops, prevents or reduces the frequency, rate or extent of myeloid cell differentiation.
regulation of macrophage differentiation
Any process that modulates the frequency, rate or extent of macrophage differentiation.
negative regulation of macrophage differentiation
Any process that stops, prevents or reduces the frequency, rate or extent of macrophage differentiation.
organ development
Development of a tissue or tissues that work together to perform a specific function or functions. Development pertains to the process whose specific outcome is the progression of a structure over time, from its formation to the mature structure. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
negative regulation of biological process
Any process that stops, prevents or reduces the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
negative regulation of cellular process
Any process that stops, prevents or reduces the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
hemopoietic or lymphoid organ development
The process whose specific outcome is the progression of any organ involved in hemopoiesis or lymphoid cell activation over time, from its formation to the mature structure. Such development includes differentiation of resident cell types (stromal cells) and of migratory cell types dependent on the unique microenvironment afforded by the organ for their proper differentiation.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
anatomical structure development
The biological process whose specific outcome is the progression of an anatomical structure from an initial condition to its mature state. This process begins with the formation of the structure and ends with the mature structure, whatever form that may be including its natural destruction. An anatomical structure is any biological entity that occupies space and is distinguished from its surroundings. Anatomical structures can be macroscopic such as a carpel, or microscopic such as an acrosome.
cellular developmental process
A biological process whose specific outcome is the progression of a cell over time from an initial condition to a later condition.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of developmental process
Any process that modulates the frequency, rate or extent of development, the biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote, or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
negative regulation of developmental process
Any process that stops, prevents or reduces the rate or extent of development, the biological process whose specific outcome is the progression of an organism over time from an initial condition (e.g. a zygote, or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
regulation of multicellular organismal process
Any process that modulates the frequency, rate or extent of a multicellular organismal process, the processes pertinent to the function of a multicellular organism above the cellular level; includes the integrated processes of tissues and organs.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
all
This term is the most general term possible
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
cellular developmental process
A biological process whose specific outcome is the progression of a cell over time from an initial condition to a later condition.
negative regulation of cellular process
Any process that stops, prevents or reduces the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
negative regulation of developmental process
Any process that stops, prevents or reduces the rate or extent of development, the biological process whose specific outcome is the progression of an organism over time from an initial condition (e.g. a zygote, or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
regulation of immune system process
Any process that modulates the frequency, rate, or extent of an immune system process.
negative regulation of biological process
Any process that stops, prevents or reduces the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of developmental process
Any process that modulates the frequency, rate or extent of development, the biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote, or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of multicellular organismal process
Any process that modulates the frequency, rate or extent of a multicellular organismal process, the processes pertinent to the function of a multicellular organism above the cellular level; includes the integrated processes of tissues and organs.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of cell proliferation
Any process that modulates the frequency, rate or extent of cell proliferation.
negative regulation of cellular process
Any process that stops, prevents or reduces the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of myeloid cell differentiation
Any process that modulates the frequency, rate or extent of myeloid cell differentiation.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
regulation of cell differentiation
Any process that modulates the frequency, rate or extent of cell differentiation, the process whereby relatively unspecialized cells acquire specialized structural and functional features.
negative regulation of developmental process
Any process that stops, prevents or reduces the rate or extent of development, the biological process whose specific outcome is the progression of an organism over time from an initial condition (e.g. a zygote, or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
negative regulation of cell differentiation
Any process that stops, prevents or reduces the frequency, rate or extent of cell differentiation.
negative regulation of myeloid cell differentiation
Any process that stops, prevents or reduces the frequency, rate or extent of myeloid cell differentiation.
regulation of cell differentiation
Any process that modulates the frequency, rate or extent of cell differentiation, the process whereby relatively unspecialized cells acquire specialized structural and functional features.
negative regulation of cell differentiation
Any process that stops, prevents or reduces the frequency, rate or extent of cell differentiation.
negative regulation of cell differentiation
Any process that stops, prevents or reduces the frequency, rate or extent of cell differentiation.
regulation of myeloid cell differentiation
Any process that modulates the frequency, rate or extent of myeloid cell differentiation.
immune system development
The process whose specific outcome is the progression of an organismal system whose objective is to provide calibrated responses by an organism to a potential internal or invasive threat, over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
organ development
Development of a tissue or tissues that work together to perform a specific function or functions. Development pertains to the process whose specific outcome is the progression of a structure over time, from its formation to the mature structure. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
hemopoietic or lymphoid organ development
The process whose specific outcome is the progression of any organ involved in hemopoiesis or lymphoid cell activation over time, from its formation to the mature structure. Such development includes differentiation of resident cell types (stromal cells) and of migratory cell types dependent on the unique microenvironment afforded by the organ for their proper differentiation.
leukocyte differentiation
The process whereby a relatively unspecialized hemopoietic precursor cell acquires the specialized features of a plasmacytoid dendritic cell or any cell of the myeloid leukocyte or lymphocyte lineages.
myeloid cell differentiation
The process whereby a relatively unspecialized myeloid precursor cell acquires the specialized features of any cell of the myeloid leukocyte, megakaryocyte, thrombocyte, or erythrocyte lineages.
negative regulation of myeloid leukocyte differentiation
Any process that stops, prevents, or reduces the frequency, rate, or extent of myeloid leukocyte differentiation.
myeloid leukocyte differentiation
The process whereby a relatively unspecialized myeloid precursor cell acquires the specialized features of any cell of the myeloid leukocyte lineage.
regulation of myeloid cell differentiation
Any process that modulates the frequency, rate or extent of myeloid cell differentiation.
negative regulation of myeloid cell differentiation
Any process that stops, prevents or reduces the frequency, rate or extent of myeloid cell differentiation.
negative regulation of macrophage differentiation
Any process that stops, prevents or reduces the frequency, rate or extent of macrophage differentiation.
regulation of myeloid leukocyte differentiation
Any process that modulates the frequency, rate, or extent of myeloid leukocyte differentiation.
negative regulation of myeloid leukocyte differentiation
Any process that stops, prevents, or reduces the frequency, rate, or extent of myeloid leukocyte differentiation.
regulation of macrophage differentiation
Any process that modulates the frequency, rate or extent of macrophage differentiation.
negative regulation of macrophage differentiation
Any process that stops, prevents or reduces the frequency, rate or extent of macrophage differentiation.

plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.

| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 04080 | 1.817e-02 | 4.841 | 14 | 83 | Neuroactive ligand-receptor interaction |
ABCA8ATP-binding cassette, sub-family A (ABC1), member 8 (204719_at), score: 0.59 ABCE1ATP-binding cassette, sub-family E (OABP), member 1 (201873_s_at), score: -0.52 ABHD5abhydrolase domain containing 5 (218739_at), score: -0.75 ABLIM3actin binding LIM protein family, member 3 (205730_s_at), score: 0.58 ACP6acid phosphatase 6, lysophosphatidic (218795_at), score: 0.58 ACSL4acyl-CoA synthetase long-chain family member 4 (202422_s_at), score: -0.65 ACSS3acyl-CoA synthetase short-chain family member 3 (219616_at), score: 0.72 ACTC1actin, alpha, cardiac muscle 1 (205132_at), score: -0.75 ACTR3BARP3 actin-related protein 3 homolog B (yeast) (218868_at), score: -0.85 ADAadenosine deaminase (204639_at), score: -0.58 ADAM19ADAM metallopeptidase domain 19 (meltrin beta) (209765_at), score: -0.85 ADARadenosine deaminase, RNA-specific (201786_s_at), score: 0.84 ADH1Balcohol dehydrogenase 1B (class I), beta polypeptide (209612_s_at), score: 0.68 ADORA2Badenosine A2b receptor (205891_at), score: 0.94 AGBL5ATP/GTP binding protein-like 5 (218480_at), score: 0.61 AGRNagrin (217419_x_at), score: 0.57 AHI1Abelson helper integration site 1 (221569_at), score: -0.81 AHNAKAHNAK nucleoprotein (211986_at), score: 0.58 AIM1absent in melanoma 1 (212543_at), score: 0.97 AJAP1adherens junctions associated protein 1 (206460_at), score: -0.58 AKR1B10aldo-keto reductase family 1, member B10 (aldose reductase) (206561_s_at), score: 0.69 AKR1C3aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II) (209160_at), score: 0.78 ALDH3A2aldehyde dehydrogenase 3 family, member A2 (202053_s_at), score: 0.69 ANGPTL2angiopoietin-like 2 (213001_at), score: 0.86 ANK2ankyrin 2, neuronal (202920_at), score: 0.6 ANXA10annexin A10 (210143_at), score: -0.8 ANXA8L2annexin A8-like 2 (203074_at), score: -0.52 APBA1amyloid beta (A4) precursor protein-binding, family A, member 1 (206679_at), score: -0.55 APOBEC3Fapolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3F (214995_s_at), score: 0.69 APOBEC3Gapolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3G (204205_at), score: 0.71 APOL3apolipoprotein L, 3 (221087_s_at), score: 0.71 APOL6apolipoprotein L, 6 (219716_at), score: 0.83 ARHGAP6Rho GTPase activating protein 6 (206167_s_at), score: 0.6 ARHGEF2rho/rac guanine nucleotide exchange factor (GEF) 2 (209435_s_at), score: 0.64 ARHGEF3Rho guanine nucleotide exchange factor (GEF) 3 (218501_at), score: 0.66 ARL4CADP-ribosylation factor-like 4C (202207_at), score: 0.71 ARMC9armadillo repeat containing 9 (219637_at), score: 0.77 ARSJarylsulfatase family, member J (219973_at), score: -0.55 ASAP3ArfGAP with SH3 domain, ankyrin repeat and PH domain 3 (219103_at), score: 0.76 ASPHD1aspartate beta-hydroxylase domain containing 1 (214993_at), score: -0.57 ATP2C1ATPase, Ca++ transporting, type 2C, member 1 (211137_s_at), score: -0.62 ATP6V0BATPase, H+ transporting, lysosomal 21kDa, V0 subunit b (200078_s_at), score: -0.71 ATXN1ataxin 1 (203231_s_at), score: 0.6 B9D1B9 protein domain 1 (210534_s_at), score: 0.71 BAT1HLA-B associated transcript 1 (200041_s_at), score: 0.59 BAZ1Abromodomain adjacent to zinc finger domain, 1A (217985_s_at), score: -0.53 BCAS4breast carcinoma amplified sequence 4 (220588_at), score: -0.52 BDH23-hydroxybutyrate dehydrogenase, type 2 (218285_s_at), score: 0.59 BDKRB2bradykinin receptor B2 (205870_at), score: 0.66 BEX4brain expressed, X-linked 4 (215440_s_at), score: 0.57 BIN1bridging integrator 1 (210201_x_at), score: 0.62 BMP6bone morphogenetic protein 6 (206176_at), score: -0.9 BNC2basonuclin 2 (220272_at), score: 0.63 BTBD2BTB (POZ) domain containing 2 (207722_s_at), score: 0.65 BTG1B-cell translocation gene 1, anti-proliferative (200920_s_at), score: 0.59 BTG2BTG family, member 2 (201236_s_at), score: 0.72 C11orf30chromosome 11 open reading frame 30 (219012_s_at), score: -0.52 C11orf41chromosome 11 open reading frame 41 (214772_at), score: -0.64 C11orf75chromosome 11 open reading frame 75 (219806_s_at), score: -0.57 C13orf15chromosome 13 open reading frame 15 (218723_s_at), score: 0.73 C14orf139chromosome 14 open reading frame 139 (219563_at), score: 0.64 C14orf159chromosome 14 open reading frame 159 (218298_s_at), score: 0.64 C14orf45chromosome 14 open reading frame 45 (220173_at), score: -0.58 C14orf94chromosome 14 open reading frame 94 (218383_at), score: 0.57 C16orf45chromosome 16 open reading frame 45 (212736_at), score: 0.57 C1orf107chromosome 1 open reading frame 107 (214193_s_at), score: -0.78 C1orf9chromosome 1 open reading frame 9 (203429_s_at), score: -0.53 C20orf117chromosome 20 open reading frame 117 (207711_at), score: 0.63 C2CD2C2 calcium-dependent domain containing 2 (212875_s_at), score: -0.62 C2CD2LC2CD2-like (204757_s_at), score: -0.59 C3orf64chromosome 3 open reading frame 64 (221935_s_at), score: -0.62 C4orf43chromosome 4 open reading frame 43 (218513_at), score: -0.53 C5orf23chromosome 5 open reading frame 23 (219054_at), score: -0.69 CALB2calbindin 2 (205428_s_at), score: -0.9 CALCOCO1calcium binding and coiled-coil domain 1 (209002_s_at), score: 0.82 CAMLGcalcium modulating ligand (203538_at), score: 0.7 CASC3cancer susceptibility candidate 3 (207842_s_at), score: 0.6 CCDC28Acoiled-coil domain containing 28A (209479_at), score: 0.64 CCNG1cyclin G1 (208796_s_at), score: 0.65 CCNG2cyclin G2 (211559_s_at), score: 0.58 CD55CD55 molecule, decay accelerating factor for complement (Cromer blood group) (201925_s_at), score: -0.61 CDC25Bcell division cycle 25 homolog B (S. pombe) (201853_s_at), score: 0.71 CDC2L6cell division cycle 2-like 6 (CDK8-like) (212899_at), score: 0.68 CDC42EP4CDC42 effector protein (Rho GTPase binding) 4 (214721_x_at), score: 0.58 CDCP1CUB domain containing protein 1 (218451_at), score: -0.66 CDH2cadherin 2, type 1, N-cadherin (neuronal) (203441_s_at), score: -0.56 CDH4cadherin 4, type 1, R-cadherin (retinal) (206866_at), score: -0.68 CDH6cadherin 6, type 2, K-cadherin (fetal kidney) (205532_s_at), score: -0.54 CDKN1Ccyclin-dependent kinase inhibitor 1C (p57, Kip2) (213348_at), score: 0.71 CEP68centrosomal protein 68kDa (212677_s_at), score: 0.7 CEP70centrosomal protein 70kDa (219036_at), score: 0.58 CHRNA10cholinergic receptor, nicotinic, alpha 10 (220210_at), score: -0.54 CHRNB2cholinergic receptor, nicotinic, beta 2 (neuronal) (206635_at), score: -0.54 CHUKconserved helix-loop-helix ubiquitous kinase (209666_s_at), score: -0.6 CIZ1CDKN1A interacting zinc finger protein 1 (205516_x_at), score: 0.61 CLIP3CAP-GLY domain containing linker protein 3 (212358_at), score: 0.73 CMPK1cytidine monophosphate (UMP-CMP) kinase 1, cytosolic (217870_s_at), score: -0.62 COL18A1collagen, type XVIII, alpha 1 (209081_s_at), score: 0.64 CRBNcereblon (218142_s_at), score: 0.77 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: 0.89 CSF1colony stimulating factor 1 (macrophage) (209716_at), score: 0.6 CSGALNACT2chondroitin sulfate N-acetylgalactosaminyltransferase 2 (222235_s_at), score: -0.54 CST6cystatin E/M (206595_at), score: -0.59 CTDSP2CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 (203445_s_at), score: 0.84 CUX1cut-like homeobox 1 (214743_at), score: -0.65 CXCL6chemokine (C-X-C motif) ligand 6 (granulocyte chemotactic protein 2) (206336_at), score: -0.52 DAAM2dishevelled associated activator of morphogenesis 2 (212793_at), score: 0.64 DAPK1death-associated protein kinase 1 (203139_at), score: 0.71 DCKdeoxycytidine kinase (203302_at), score: -0.64 DCLK1doublecortin-like kinase 1 (205399_at), score: 0.61 DCP2DCP2 decapping enzyme homolog (S. cerevisiae) (212919_at), score: -0.53 DDB2damage-specific DNA binding protein 2, 48kDa (203409_at), score: 0.71 DDX58DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 (218943_s_at), score: 0.73 DDX60DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 (218986_s_at), score: 0.75 DENND2ADENN/MADD domain containing 2A (53991_at), score: 0.83 DENND5BDENN/MADD domain containing 5B (215058_at), score: -0.64 DEPDC6DEP domain containing 6 (218858_at), score: 0.82 DERL2Der1-like domain family, member 2 (218333_at), score: -0.54 DIRAS3DIRAS family, GTP-binding RAS-like 3 (215506_s_at), score: -0.8 DLSTPdihydrolipoamide S-succinyltransferase pseudogene (E2 component of 2-oxo-glutarate complex) (215210_s_at), score: -0.53 DMPKdystrophia myotonica-protein kinase (37996_s_at), score: 0.83 DNAJB9DnaJ (Hsp40) homolog, subfamily B, member 9 (202843_at), score: -0.92 DNAJC1DnaJ (Hsp40) homolog, subfamily C, member 1 (218409_s_at), score: -0.56 DOCK10dedicator of cytokinesis 10 (219279_at), score: -0.68 DSPdesmoplakin (200606_at), score: -0.54 DTX4deltex homolog 4 (Drosophila) (212611_at), score: 0.64 DUSP14dual specificity phosphatase 14 (203367_at), score: -0.78 DUSP4dual specificity phosphatase 4 (204014_at), score: -0.68 DZIP3DAZ interacting protein 3, zinc finger (213186_at), score: 0.8 EDEM1ER degradation enhancer, mannosidase alpha-like 1 (203279_at), score: -0.79 EFHC1EF-hand domain (C-terminal) containing 1 (219833_s_at), score: 0.59 EFNA5ephrin-A5 (214036_at), score: 0.65 EHMT2euchromatic histone-lysine N-methyltransferase 2 (202326_at), score: 0.6 EIF4Eeukaryotic translation initiation factor 4E (201436_at), score: -0.69 EIF5Beukaryotic translation initiation factor 5B (214314_s_at), score: -0.6 ELTD1EGF, latrophilin and seven transmembrane domain containing 1 (219134_at), score: -0.63 EML2echinoderm microtubule associated protein like 2 (204398_s_at), score: -0.52 ENOX1ecto-NOX disulfide-thiol exchanger 1 (219501_at), score: 0.74 ENTPD1ectonucleoside triphosphate diphosphohydrolase 1 (209473_at), score: -0.63 EPHA4EPH receptor A4 (206114_at), score: -0.55 EPHX1epoxide hydrolase 1, microsomal (xenobiotic) (202017_at), score: 0.67 ESM1endothelial cell-specific molecule 1 (208394_x_at), score: -0.89 ESRRAestrogen-related receptor alpha (1487_at), score: -0.68 EXD3exonuclease 3'-5' domain containing 3 (220838_at), score: -0.52 FAM102Afamily with sequence similarity 102, member A (212400_at), score: 0.62 FAM155Afamily with sequence similarity 155, member A (214825_at), score: -0.74 FAM18Bfamily with sequence similarity 18, member B (218446_s_at), score: -0.6 FARSAphenylalanyl-tRNA synthetase, alpha subunit (216602_s_at), score: -0.53 FBXL14F-box and leucine-rich repeat protein 14 (213145_at), score: 0.75 FBXL7F-box and leucine-rich repeat protein 7 (213249_at), score: 0.68 FERMT1fermitin family homolog 1 (Drosophila) (218796_at), score: -0.69 FGF7fibroblast growth factor 7 (keratinocyte growth factor) (205782_at), score: -0.57 FGGYFGGY carbohydrate kinase domain containing (219718_at), score: 0.6 FICDFIC domain containing (219910_at), score: -0.74 FLJ12529pre-mRNA cleavage factor I, 59 kDa subunit (217866_at), score: 0.61 FLNCfilamin C, gamma (actin binding protein 280) (207876_s_at), score: -0.61 FLT3LGfms-related tyrosine kinase 3 ligand (210607_at), score: 0.7 FNBP1formin binding protein 1 (212288_at), score: 0.88 FNDC3Afibronectin type III domain containing 3A (202304_at), score: -0.63 FOLR3folate receptor 3 (gamma) (206371_at), score: -0.69 FOSL2FOS-like antigen 2 (218880_at), score: 0.61 FOXK2forkhead box K2 (203064_s_at), score: 0.58 FSTL3follistatin-like 3 (secreted glycoprotein) (203592_s_at), score: -0.76 FTSJ1FtsJ homolog 1 (E. coli) (213937_s_at), score: -0.53 FYNFYN oncogene related to SRC, FGR, YES (210105_s_at), score: 0.75 GABBR2gamma-aminobutyric acid (GABA) B receptor, 2 (209990_s_at), score: -0.77 GABREgamma-aminobutyric acid (GABA) A receptor, epsilon (204537_s_at), score: 0.59 GALgalanin prepropeptide (214240_at), score: -0.69 GBP2guanylate binding protein 2, interferon-inducible (202748_at), score: 0.6 GFOD1glucose-fructose oxidoreductase domain containing 1 (219821_s_at), score: -0.78 GHRgrowth hormone receptor (205498_at), score: -0.58 GKglycerol kinase (207387_s_at), score: -0.85 GK3Pglycerol kinase 3 pseudogene (215966_x_at), score: -0.99 GLTSCR2glioma tumor suppressor candidate region gene 2 (217807_s_at), score: 0.7 GMCL1germ cell-less homolog 1 (Drosophila) (218458_at), score: -0.69 GMPPBGDP-mannose pyrophosphorylase B (219920_s_at), score: -0.57 GMPRguanosine monophosphate reductase (204187_at), score: 0.83 GOLT1Bgolgi transport 1 homolog B (S. cerevisiae) (218193_s_at), score: -0.71 GPR177G protein-coupled receptor 177 (221958_s_at), score: -0.55 GPRC5BG protein-coupled receptor, family C, group 5, member B (203632_s_at), score: 0.63 GPSM2G-protein signaling modulator 2 (AGS3-like, C. elegans) (221922_at), score: 0.67 GRAMD1BGRAM domain containing 1B (212906_at), score: -0.66 GRB14growth factor receptor-bound protein 14 (206204_at), score: -0.55 GRSF1G-rich RNA sequence binding factor 1 (201501_s_at), score: -0.66 GTF2A1Lgeneral transcription factor IIA, 1-like (213413_at), score: 1 GUCY1B3guanylate cyclase 1, soluble, beta 3 (203817_at), score: 0.66 H1F0H1 histone family, member 0 (208886_at), score: 0.77 H1FXH1 histone family, member X (204805_s_at), score: 0.65 HAS2hyaluronan synthase 2 (206432_at), score: -0.56 HDGFhepatoma-derived growth factor (high-mobility group protein 1-like) (200896_x_at), score: 0.78 HERC6hect domain and RLD 6 (219352_at), score: 0.64 HGFhepatocyte growth factor (hepapoietin A; scatter factor) (209960_at), score: 0.62 HHIPL2HHIP-like 2 (220283_at), score: -0.62 HISPPD1histidine acid phosphatase domain containing 1 (203253_s_at), score: -0.58 HIST1H4Ghistone cluster 1, H4g (208551_at), score: -0.53 HIST3H2Ahistone cluster 3, H2a (221582_at), score: 0.61 HLA-DQB1major histocompatibility complex, class II, DQ beta 1 (211654_x_at), score: -0.61 HMG20Bhigh-mobility group 20B (210719_s_at), score: 0.59 HOMER2homer homolog 2 (Drosophila) (217080_s_at), score: -0.54 HS3ST3A1heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1 (219985_at), score: -0.52 HSD17B6hydroxysteroid (17-beta) dehydrogenase 6 homolog (mouse) (37512_at), score: -0.54 HSD17B7hydroxysteroid (17-beta) dehydrogenase 7 (220081_x_at), score: 0.65 HSPA12Aheat shock 70kDa protein 12A (214434_at), score: 0.71 HSPA4Lheat shock 70kDa protein 4-like (205543_at), score: 0.73 HTR2A5-hydroxytryptamine (serotonin) receptor 2A (207135_at), score: -0.7 HTRA1HtrA serine peptidase 1 (201185_at), score: 0.58 ICAM3intercellular adhesion molecule 3 (204949_at), score: -0.6 ID4inhibitor of DNA binding 4, dominant negative helix-loop-helix protein (209291_at), score: -0.57 IFI16interferon, gamma-inducible protein 16 (208965_s_at), score: 0.62 IFI35interferon-induced protein 35 (209417_s_at), score: 0.65 IFI44interferon-induced protein 44 (214453_s_at), score: 0.65 IFIH1interferon induced with helicase C domain 1 (219209_at), score: 0.93 IFIT3interferon-induced protein with tetratricopeptide repeats 3 (204747_at), score: 0.77 IL11interleukin 11 (206924_at), score: -0.74 IL13RA2interleukin 13 receptor, alpha 2 (206172_at), score: -0.7 IL24interleukin 24 (206569_at), score: -0.55 IL33interleukin 33 (209821_at), score: -0.69 INE1inactivation escape 1 (non-protein coding) (207252_at), score: -0.58 INHBAinhibin, beta A (210511_s_at), score: -0.62 IRF1interferon regulatory factor 1 (202531_at), score: 0.92 IRF2interferon regulatory factor 2 (203275_at), score: 0.74 IRF9interferon regulatory factor 9 (203882_at), score: 0.66 ITGAVintegrin, alpha V (vitronectin receptor, alpha polypeptide, antigen CD51) (202351_at), score: -0.55 JUPjunction plakoglobin (201015_s_at), score: 0.6 KAL1Kallmann syndrome 1 sequence (205206_at), score: -0.89 KCNG1potassium voltage-gated channel, subfamily G, member 1 (214595_at), score: -0.64 KCNK1potassium channel, subfamily K, member 1 (204679_at), score: -0.54 KIAA0556KIAA0556 (213185_at), score: 0.57 KIAA0562KIAA0562 (204075_s_at), score: -0.53 KIAA1305KIAA1305 (220911_s_at), score: 0.81 KIAA1462KIAA1462 (213316_at), score: 0.66 KIF13Bkinesin family member 13B (202962_at), score: 0.58 KLHDC2kelch domain containing 2 (217906_at), score: 0.65 KLHL23kelch-like 23 (Drosophila) (213610_s_at), score: -0.64 LARP4La ribonucleoprotein domain family, member 4 (214155_s_at), score: -0.6 LDLRlow density lipoprotein receptor (202068_s_at), score: 0.61 LEPREL1leprecan-like 1 (218717_s_at), score: -0.66 LETMD1LETM1 domain containing 1 (207170_s_at), score: 0.62 LIMK2LIM domain kinase 2 (202193_at), score: -0.7 LIN7Blin-7 homolog B (C. elegans) (219760_at), score: -0.59 LLGL2lethal giant larvae homolog 2 (Drosophila) (203713_s_at), score: -0.53 LOC100132540similar to LOC339047 protein (214870_x_at), score: 0.87 LOC100133105hypothetical protein LOC100133105 (214237_x_at), score: -0.76 LOC339047hypothetical protein LOC339047 (221501_x_at), score: 0.89 LOC399491LOC399491 protein (214035_x_at), score: 0.85 LOC440434hypothetical protein FLJ11822 (215090_x_at), score: 0.71 LRP8low density lipoprotein receptor-related protein 8, apolipoprotein e receptor (205282_at), score: -0.91 LRRC49leucine rich repeat containing 49 (219338_s_at), score: 0.61 LRRFIP1leucine rich repeat (in FLII) interacting protein 1 (201861_s_at), score: -0.55 LRRN3leucine rich repeat neuronal 3 (209841_s_at), score: -0.51 LY75lymphocyte antigen 75 (205668_at), score: 0.64 LYRM1LYR motif containing 1 (203897_at), score: 0.63 MAN1C1mannosidase, alpha, class 1C, member 1 (218918_at), score: 0.9 MAP2K5mitogen-activated protein kinase kinase 5 (211370_s_at), score: 0.74 MAPRE2microtubule-associated protein, RP/EB family, member 2 (202501_at), score: 0.61 MBOAT2membrane bound O-acyltransferase domain containing 2 (213288_at), score: -0.71 MBPmyelin basic protein (210136_at), score: 0.6 MC4Rmelanocortin 4 receptor (221467_at), score: -0.62 MDM2Mdm2 p53 binding protein homolog (mouse) (205386_s_at), score: 0.63 MFAP3Lmicrofibrillar-associated protein 3-like (205442_at), score: -0.8 MGC87042similar to Six transmembrane epithelial antigen of prostate (217553_at), score: -0.62 MINK1misshapen-like kinase 1 (zebrafish) (214246_x_at), score: 0.57 MIS12MIS12, MIND kinetochore complex component, homolog (yeast) (221559_s_at), score: -0.53 MKNK2MAP kinase interacting serine/threonine kinase 2 (218205_s_at), score: 0.72 MMP10matrix metallopeptidase 10 (stromelysin 2) (205680_at), score: -0.92 MMP16matrix metallopeptidase 16 (membrane-inserted) (207012_at), score: -0.78 MPP1membrane protein, palmitoylated 1, 55kDa (202974_at), score: 0.57 MRASmuscle RAS oncogene homolog (206538_at), score: -0.65 MREGmelanoregulin (219648_at), score: -0.62 MSTO1misato homolog 1 (Drosophila) (218296_x_at), score: -0.8 MSX2msh homeobox 2 (210319_x_at), score: 0.64 MT1Fmetallothionein 1F (217165_x_at), score: -0.56 MT1Gmetallothionein 1G (204745_x_at), score: -0.56 MT1Mmetallothionein 1M (217546_at), score: -0.54 MT1Xmetallothionein 1X (204326_x_at), score: -0.68 MT2Ametallothionein 2A (212185_x_at), score: -0.54 MTAPmethylthioadenosine phosphorylase (211363_s_at), score: -0.65 MX1myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) (202086_at), score: 0.69 MX2myxovirus (influenza virus) resistance 2 (mouse) (204994_at), score: 0.82 MXD4MAX dimerization protein 4 (210778_s_at), score: 0.6 MYBBP1AMYB binding protein (P160) 1a (219098_at), score: -0.63 MYL4myosin, light chain 4, alkali; atrial, embryonic (210088_x_at), score: -0.6 MYO10myosin X (201976_s_at), score: -0.67 NCALDneurocalcin delta (211685_s_at), score: -0.94 NEDD4neural precursor cell expressed, developmentally down-regulated 4 (213012_at), score: -0.59 NEFMneurofilament, medium polypeptide (205113_at), score: -0.93 NF1neurofibromin 1 (211094_s_at), score: 0.68 NFKBIAnuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha (201502_s_at), score: 0.73 NMIN-myc (and STAT) interactor (203964_at), score: 0.87 NOX4NADPH oxidase 4 (219773_at), score: -0.74 NPnucleoside phosphorylase (201695_s_at), score: -0.66 NPC2Niemann-Pick disease, type C2 (200701_at), score: 0.67 NPIPnuclear pore complex interacting protein (204538_x_at), score: 0.91 NPR2natriuretic peptide receptor B/guanylate cyclase B (atrionatriuretic peptide receptor B) (204310_s_at), score: 0.64 NPR3natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) (219789_at), score: -0.61 NPTX1neuronal pentraxin I (204684_at), score: -0.74 NR1H3nuclear receptor subfamily 1, group H, member 3 (203920_at), score: 0.68 NR5A2nuclear receptor subfamily 5, group A, member 2 (208343_s_at), score: -0.64 NTMneurotrimin (222020_s_at), score: -0.83 OAS12',5'-oligoadenylate synthetase 1, 40/46kDa (205552_s_at), score: 0.73 OAS22'-5'-oligoadenylate synthetase 2, 69/71kDa (204972_at), score: 0.96 OAS32'-5'-oligoadenylate synthetase 3, 100kDa (218400_at), score: 0.79 OLFML2Bolfactomedin-like 2B (213125_at), score: 0.69 OPCMLopioid binding protein/cell adhesion molecule-like (214111_at), score: -0.89 ORAI3ORAI calcium release-activated calcium modulator 3 (221864_at), score: 0.74 OSR2odd-skipped related 2 (Drosophila) (213568_at), score: 0.58 OSTM1osteopetrosis associated transmembrane protein 1 (218196_at), score: -0.65 OXA1Loxidase (cytochrome c) assembly 1-like (208717_at), score: 0.59 P2RX5purinergic receptor P2X, ligand-gated ion channel, 5 (210448_s_at), score: -0.82 P2RY5purinergic receptor P2Y, G-protein coupled, 5 (218589_at), score: 0.75 PARNpoly(A)-specific ribonuclease (deadenylation nuclease) (203905_at), score: 0.58 PARP12poly (ADP-ribose) polymerase family, member 12 (218543_s_at), score: 0.61 PCDH9protocadherin 9 (219737_s_at), score: -0.99 PCYT2phosphate cytidylyltransferase 2, ethanolamine (209577_at), score: 0.69 PDCD1LG2programmed cell death 1 ligand 2 (220049_s_at), score: -0.71 PDE5Aphosphodiesterase 5A, cGMP-specific (206757_at), score: 0.87 PDE8Aphosphodiesterase 8A (212522_at), score: -0.58 PDGFRAplatelet-derived growth factor receptor, alpha polypeptide (203131_at), score: 0.65 PDGFRBplatelet-derived growth factor receptor, beta polypeptide (202273_at), score: 0.6 PDPK13-phosphoinositide dependent protein kinase-1 (204524_at), score: -0.58 PECRperoxisomal trans-2-enoyl-CoA reductase (221142_s_at), score: 0.58 PENKproenkephalin (213791_at), score: -0.52 PFN2profilin 2 (204992_s_at), score: 0.64 PHC1polyhomeotic homolog 1 (Drosophila) (218338_at), score: 0.71 PIGHphosphatidylinositol glycan anchor biosynthesis, class H (209625_at), score: -0.59 PIGLphosphatidylinositol glycan anchor biosynthesis, class L (213889_at), score: 0.73 PITX1paired-like homeodomain 1 (208502_s_at), score: 0.74 PKIGprotein kinase (cAMP-dependent, catalytic) inhibitor gamma (202732_at), score: 0.65 PLA2G4Cphospholipase A2, group IVC (cytosolic, calcium-independent) (209785_s_at), score: 0.59 PLAUplasminogen activator, urokinase (211668_s_at), score: 0.62 PLEK2pleckstrin 2 (218644_at), score: -0.53 PLEKHA5pleckstrin homology domain containing, family A member 5 (220952_s_at), score: 0.67 PLSCR1phospholipid scramblase 1 (202446_s_at), score: 0.59 PMP22peripheral myelin protein 22 (210139_s_at), score: 0.61 POLR3Kpolymerase (RNA) III (DNA directed) polypeptide K, 12.3 kDa (218866_s_at), score: -0.54 PPARGperoxisome proliferator-activated receptor gamma (208510_s_at), score: 0.82 PPATphosphoribosyl pyrophosphate amidotransferase (209433_s_at), score: -0.66 PPP2R1Bprotein phosphatase 2 (formerly 2A), regulatory subunit A, beta isoform (202884_s_at), score: -0.61 PPP3R1protein phosphatase 3 (formerly 2B), regulatory subunit B, alpha isoform (204507_s_at), score: -0.61 PRKCHprotein kinase C, eta (218764_at), score: 0.61 PRSS2protease, serine, 2 (trypsin 2) (205402_x_at), score: -0.69 PRSS3protease, serine, 3 (207463_x_at), score: -0.57 PSMB8proteasome (prosome, macropain) subunit, beta type, 8 (large multifunctional peptidase 7) (209040_s_at), score: 0.78 PSMB9proteasome (prosome, macropain) subunit, beta type, 9 (large multifunctional peptidase 2) (204279_at), score: 0.61 PTGER4prostaglandin E receptor 4 (subtype EP4) (204897_at), score: 0.59 PTGS1prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) (215813_s_at), score: -0.66 PTGS2prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) (204748_at), score: -0.73 PTHLHparathyroid hormone-like hormone (211756_at), score: -0.83 PTMAP7prothymosin, alpha pseudogene 7 (208549_x_at), score: -0.53 PTP4A1protein tyrosine phosphatase type IVA, member 1 (200730_s_at), score: -0.67 PTPRBprotein tyrosine phosphatase, receptor type, B (205846_at), score: -0.68 PTPROprotein tyrosine phosphatase, receptor type, O (211600_at), score: 0.69 PVRL3poliovirus receptor-related 3 (213325_at), score: -0.55 PYGLphosphorylase, glycogen, liver (202990_at), score: 0.62 RAB1ARAB1A, member RAS oncogene family (213440_at), score: -0.77 RAB21RAB21, member RAS oncogene family (203885_at), score: -0.55 RAP1GAPRAP1 GTPase activating protein (203911_at), score: -0.55 RASIP1Ras interacting protein 1 (220027_s_at), score: -0.54 RASSF2Ras association (RalGDS/AF-6) domain family member 2 (203185_at), score: 0.73 RBAKRB-associated KRAB zinc finger (219214_s_at), score: -0.52 RBM4BRNA binding motif protein 4B (209497_s_at), score: 0.61 RCAN2regulator of calcineurin 2 (203498_at), score: 0.67 REREarginine-glutamic acid dipeptide (RE) repeats (200940_s_at), score: 0.69 RFX5regulatory factor X, 5 (influences HLA class II expression) (202963_at), score: 0.78 RGS17regulator of G-protein signaling 17 (220334_at), score: 0.67 RGS20regulator of G-protein signaling 20 (210138_at), score: -0.58 RHBDF1rhomboid 5 homolog 1 (Drosophila) (218686_s_at), score: 0.61 RINT1RAD50 interactor 1 (218598_at), score: -0.53 RNF220ring finger protein 220 (219988_s_at), score: 0.68 RNF44ring finger protein 44 (203286_at), score: 0.71 ROR2receptor tyrosine kinase-like orphan receptor 2 (205578_at), score: 0.81 RP11-345P4.4similar to solute carrier family 35, member E2 (217122_s_at), score: 0.69 RP4-691N24.1ninein-like (207705_s_at), score: 0.65 RPL13Aribosomal protein L13a (200715_x_at), score: 0.64 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: -0.58 RPL23ribosomal protein L23 (214744_s_at), score: -0.6 S1PR1sphingosine-1-phosphate receptor 1 (204642_at), score: -0.73 SAV1salvador homolog 1 (Drosophila) (218276_s_at), score: 0.76 SCAMP4secretory carrier membrane protein 4 (213244_at), score: 0.67 SCG5secretogranin V (7B2 protein) (203889_at), score: -0.78 SDCBPsyndecan binding protein (syntenin) (200958_s_at), score: 0.65 SDF2L1stromal cell-derived factor 2-like 1 (218681_s_at), score: -0.73 SDPRserum deprivation response (phosphatidylserine binding protein) (218711_s_at), score: 0.58 SEC23BSec23 homolog B (S. cerevisiae) (201583_s_at), score: -0.56 SECTM1secreted and transmembrane 1 (213716_s_at), score: 0.89 SELENBP1selenium binding protein 1 (214433_s_at), score: 0.72 SELPLGselectin P ligand (209879_at), score: -0.64 SEMA3Fsema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F (35666_at), score: -0.53 SERPINB2serpin peptidase inhibitor, clade B (ovalbumin), member 2 (204614_at), score: -0.53 SERPINB9serpin peptidase inhibitor, clade B (ovalbumin), member 9 (209723_at), score: 0.66 SESN1sestrin 1 (218346_s_at), score: 0.74 SF1splicing factor 1 (208313_s_at), score: 0.61 SFRS5splicing factor, arginine/serine-rich 5 (212266_s_at), score: 0.65 SFXN1sideroflexin 1 (218392_x_at), score: -0.59 SGSHN-sulfoglucosamine sulfohydrolase (35626_at), score: 0.61 SIRT3sirtuin (silent mating type information regulation 2 homolog) 3 (S. cerevisiae) (221562_s_at), score: 0.64 SIX2SIX homeobox 2 (206510_at), score: 0.71 SLC16A1solute carrier family 16, member 1 (monocarboxylic acid transporter 1) (209900_s_at), score: -0.52 SLC22A4solute carrier family 22 (organic cation/ergothioneine transporter), member 4 (205896_at), score: -0.56 SLC25A13solute carrier family 25, member 13 (citrin) (203775_at), score: -0.63 SLC25A32solute carrier family 25, member 32 (221020_s_at), score: -0.63 SLC39A7solute carrier family 39 (zinc transporter), member 7 (202667_s_at), score: -0.56 SLC48A1solute carrier family 48 (heme transporter), member 1 (218417_s_at), score: 0.61 SLC4A4solute carrier family 4, sodium bicarbonate cotransporter, member 4 (203908_at), score: -0.84 SLC4A7solute carrier family 4, sodium bicarbonate cotransporter, member 7 (209884_s_at), score: -0.72 SLC6A11solute carrier family 6 (neurotransmitter transporter, GABA), member 11 (207048_at), score: -0.53 SLC7A6solute carrier family 7 (cationic amino acid transporter, y+ system), member 6 (203578_s_at), score: -0.54 SLC8A1solute carrier family 8 (sodium/calcium exchanger), member 1 (207053_at), score: -0.68 SLMO2slowmo homolog 2 (Drosophila) (217851_s_at), score: -0.61 SMAD3SMAD family member 3 (218284_at), score: 0.96 SMURF2SMAD specific E3 ubiquitin protein ligase 2 (205596_s_at), score: -0.56 SNAP25synaptosomal-associated protein, 25kDa (202508_s_at), score: 0.6 SNCAsynuclein, alpha (non A4 component of amyloid precursor) (204466_s_at), score: 0.64 SNNstannin (218032_at), score: 0.75 SNRNP27small nuclear ribonucleoprotein 27kDa (U4/U6.U5) (212440_at), score: -0.56 SNX27sorting nexin family member 27 (221006_s_at), score: 0.6 SPATA20spermatogenesis associated 20 (218164_at), score: 0.62 SPHK1sphingosine kinase 1 (219257_s_at), score: -0.55 SRPRBsignal recognition particle receptor, B subunit (218140_x_at), score: -0.6 SSR3signal sequence receptor, gamma (translocon-associated protein gamma) (217790_s_at), score: -0.6 SSTR1somatostatin receptor 1 (208482_at), score: -0.79 ST5suppression of tumorigenicity 5 (202440_s_at), score: 0.8 STARD5StAR-related lipid transfer (START) domain containing 5 (213820_s_at), score: 0.63 STIM1stromal interaction molecule 1 (202764_at), score: 0.65 SVILsupervillin (202565_s_at), score: 0.66 SYNJ1synaptojanin 1 (212990_at), score: -0.65 TAF13TAF13 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 18kDa (205966_at), score: -0.6 TBC1D2BTBC1 domain family, member 2B (212796_s_at), score: 0.71 TBC1D8TBC1 domain family, member 8 (with GRAM domain) (204526_s_at), score: 0.87 TCEB1transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C) (202823_at), score: -0.6 TENC1tensin like C1 domain containing phosphatase (tensin 2) (212494_at), score: 0.81 TFDP2transcription factor Dp-2 (E2F dimerization partner 2) (203588_s_at), score: 0.61 TGDSTDP-glucose 4,6-dehydratase (208249_s_at), score: -0.7 TGFAtransforming growth factor, alpha (205016_at), score: -0.71 THSD7Athrombospondin, type I, domain containing 7A (214920_at), score: 0.83 THUMPD2THUMP domain containing 2 (219248_at), score: -0.55 THYN1thymocyte nuclear protein 1 (218491_s_at), score: 0.65 TIAF1TGFB1-induced anti-apoptotic factor 1 (202039_at), score: 0.62 TICAM2toll-like receptor adaptor molecule 2 (214658_at), score: -0.66 TLR3toll-like receptor 3 (206271_at), score: 0.99 TM4SF1transmembrane 4 L six family member 1 (209386_at), score: -0.69 TM9SF3transmembrane 9 superfamily member 3 (217758_s_at), score: -0.56 TMED7transmembrane emp24 protein transport domain containing 7 (209404_s_at), score: -0.56 TMEFF1transmembrane protein with EGF-like and two follistatin-like domains 1 (205122_at), score: -0.69 TMEM104transmembrane protein 104 (220097_s_at), score: -0.65 TMEM140transmembrane protein 140 (218999_at), score: 0.68 TMEM2transmembrane protein 2 (218113_at), score: -0.84 TMEM208transmembrane protein 208 (221597_s_at), score: -0.57 TNFAIP2tumor necrosis factor, alpha-induced protein 2 (202510_s_at), score: 0.7 TNFAIP6tumor necrosis factor, alpha-induced protein 6 (206026_s_at), score: 0.64 TNS3tensin 3 (217853_at), score: 0.71 TOMM20translocase of outer mitochondrial membrane 20 homolog (yeast) (200662_s_at), score: -0.59 TP53tumor protein p53 (201746_at), score: 0.9 TP53I11tumor protein p53 inducible protein 11 (214667_s_at), score: 0.57 TRIM21tripartite motif-containing 21 (204804_at), score: 0.83 TRIM24tripartite motif-containing 24 (213301_x_at), score: -0.54 TRMT2ATRM2 tRNA methyltransferase 2 homolog A (S. cerevisiae) (91617_at), score: -0.65 TRPC4transient receptor potential cation channel, subfamily C, member 4 (220817_at), score: -0.68 TSC22D1TSC22 domain family, member 1 (215111_s_at), score: 0.6 TSPAN13tetraspanin 13 (217979_at), score: -0.83 TSPAN2tetraspanin 2 (214606_at), score: -0.52 TUBA4Btubulin, alpha 4b (pseudogene) (207490_at), score: -0.56 UAP1UDP-N-acteylglucosamine pyrophosphorylase 1 (209340_at), score: -0.63 UBE2L6ubiquitin-conjugating enzyme E2L 6 (201649_at), score: 0.84 UBXN1UBX domain protein 1 (201871_s_at), score: 0.67 UCK2uridine-cytidine kinase 2 (209825_s_at), score: -0.53 UGDHUDP-glucose dehydrogenase (203343_at), score: -0.56 UHRF1BP1LUHRF1 binding protein 1-like (213118_at), score: -0.61 ULBP2UL16 binding protein 2 (221291_at), score: -0.89 ULK1unc-51-like kinase 1 (C. elegans) (209333_at), score: 0.68 USP21ubiquitin specific peptidase 21 (218367_x_at), score: 0.61 VWA5Avon Willebrand factor A domain containing 5A (210102_at), score: 0.82 WBP2WW domain binding protein 2 (209117_at), score: 0.68 WDR4WD repeat domain 4 (221632_s_at), score: -0.66 WDR42AWD repeat domain 42A (202249_s_at), score: 0.64 WDR6WD repeat domain 6 (217734_s_at), score: 0.82 XAF1XIAP associated factor 1 (206133_at), score: 0.67 XPCxeroderma pigmentosum, complementation group C (209375_at), score: 0.6 XYLT1xylosyltransferase I (213725_x_at), score: -0.82 YDD19YDD19 protein (37079_at), score: -0.55 YES1v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 (202932_at), score: 0.68 YPEL1yippee-like 1 (Drosophila) (206063_x_at), score: -0.51 ZBTB20zinc finger and BTB domain containing 20 (205383_s_at), score: 0.72 ZDHHC7zinc finger, DHHC-type containing 7 (218606_at), score: 0.77 ZFP36L2zinc finger protein 36, C3H type-like 2 (201367_s_at), score: 0.59 ZFYVE16zinc finger, FYVE domain containing 16 (203651_at), score: -0.53 ZNF281zinc finger protein 281 (218401_s_at), score: -0.58 ZNF580zinc finger protein 580 (220748_s_at), score: 0.61 ZNF706zinc finger protein 706 (218059_at), score: 0.66
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486351.cel | 36 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690199.cel | 1 | 5 | HGPS | hgu133a | none | GM0316B |
| E-GEOD-3860-raw-cel-1561690256.cel | 6 | 5 | HGPS | hgu133a | none | GMO8398C |
| 46B.CEL | 2 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 2 |
| E-TABM-263-raw-cel-1515486331.cel | 35 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486051.cel | 21 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 2433_CNTL.CEL | 4 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| E-GEOD-3407-raw-cel-1437949938.cel | 8 | 4 | Cockayne | hgu133a | none | CSB |
| E-GEOD-3407-raw-cel-1437949704.cel | 4 | 4 | Cockayne | hgu133a | CS | eGFP |
| E-GEOD-3407-raw-cel-1437949721.cel | 5 | 4 | Cockayne | hgu133a | CS | CSB |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 8495_CNTL.CEL | 10 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| E-TABM-263-raw-cel-1515486271.cel | 32 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485791.cel | 8 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486311.cel | 34 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486291.cel | 33 | 6 | Cycle | hgu133a2 | none | Cycle 1 |