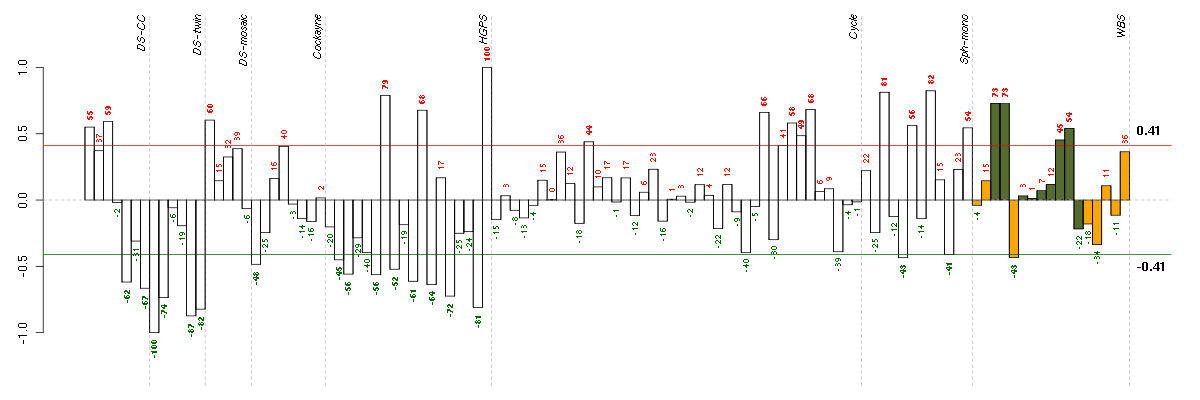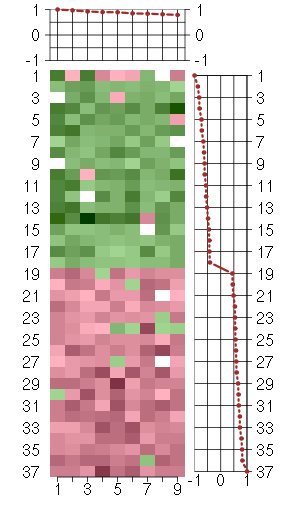

Under-expression is coded with green,
over-expression with red color.

immune system process
Any process involved in the development or functioning of the immune system, an organismal system for calibrated responses to potential internal or invasive threats.
immune response
Any immune system process that functions in the calibrated response of an organism to a potential internal or invasive threat.
humoral immune response
An immune response mediated through a body fluid.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
response to stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
all
This term is the most general term possible
immune response
Any immune system process that functions in the calibrated response of an organism to a potential internal or invasive threat.

extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
all
This term is the most general term possible

CCL2chemokine (C-C motif) ligand 2 (216598_s_at), score: 1 CFHcomplement factor H (213800_at), score: 0.9 CFHR1complement factor H-related 1 (215388_s_at), score: 0.85 COL15A1collagen, type XV, alpha 1 (203477_at), score: 0.92 PLSCR4phospholipid scramblase 4 (218901_at), score: 0.97 PROS1protein S (alpha) (207808_s_at), score: 0.81 RCAN2regulator of calcineurin 2 (203498_at), score: 0.83 THBS2thrombospondin 2 (203083_at), score: 0.79 TNFAIP8tumor necrosis factor, alpha-induced protein 8 (210260_s_at), score: 0.89
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| 1Twin.CEL | 1 | 2 | DS-twin | hgu133plus2 | Down | DS-twin 1 |
| 5CTwin.CEL | 5 | 2 | DS-twin | hgu133plus2 | Down | DS-twin 5 |
| 6Twin.CEL | 6 | 2 | DS-twin | hgu133plus2 | none | DS-twin 6 |
| E-GEOD-3860-raw-cel-1561690472.cel | 17 | 5 | HGPS | hgu133a | none | GM00038C |
| 2Twin.CEL | 2 | 2 | DS-twin | hgu133plus2 | none | DS-twin 2 |
| E-GEOD-3860-raw-cel-1561690392.cel | 14 | 5 | HGPS | hgu133a | none | GMO8398C |
| t21d 08-03.CEL | 7 | 1 | DS-CC | hgu133a | Down | DS-CC 7 |
| E-GEOD-3860-raw-cel-1561690360.cel | 12 | 5 | HGPS | hgu133a | none | GM0316B |
| t21b 08-03.CEL | 5 | 1 | DS-CC | hgu133a | Down | DS-CC 5 |
| E-GEOD-3860-raw-cel-1561690344.cel | 10 | 5 | HGPS | hgu133a | none | GM00038C |
| E-GEOD-3860-raw-cel-1561690256.cel | 6 | 5 | HGPS | hgu133a | none | GMO8398C |
| E-GEOD-3860-raw-cel-1561690223.cel | 3 | 5 | HGPS | hgu133a | none | GM00038C |
| E-GEOD-3860-raw-cel-1561690304.cel | 8 | 5 | HGPS | hgu133a | none | GMO8398C |
| E-GEOD-3407-raw-cel-1437949557.cel | 1 | 4 | Cockayne | hgu133a | CS | eGFP |
| E-GEOD-3860-raw-cel-1561690215.cel | 2 | 5 | HGPS | hgu133a | HGPS | AG11513 |
| E-GEOD-4219-raw-cel-1311956321.cel | 9 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| 4319_WBS.CEL | 5 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| E-GEOD-4219-raw-cel-1311956614.cel | 18 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-TABM-263-raw-cel-1515485851.cel | 11 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 8495_CNTL.CEL | 10 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| E-TABM-263-raw-cel-1515486311.cel | 34 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 9118_CNTL.CEL | 11 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| E-GEOD-4219-raw-cel-1311956824.cel | 24 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| ctrl a 08-03.CEL | 1 | 1 | DS-CC | hgu133a | none | DS-CC 1 |
| E-GEOD-4219-raw-cel-1311956358.cel | 10 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-TABM-263-raw-cel-1515486291.cel | 33 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| ctrl c 08-03.CEL | 3 | 1 | DS-CC | hgu133a | none | DS-CC 3 |
| 46A.CEL | 1 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 1 |
| E-TABM-263-raw-cel-1515486231.cel | 30 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690352.cel | 11 | 5 | HGPS | hgu133a | HGPS | AG11498 |
| E-TABM-263-raw-cel-1515486331.cel | 35 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 2433_CNTL.CEL | 4 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| 1104_CNTL.CEL | 3 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| E-GEOD-3860-raw-cel-1561690272.cel | 7 | 5 | HGPS | hgu133a | HGPS | AG11498 |
| E-GEOD-4219-raw-cel-1311956178.cel | 6 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956418.cel | 13 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-3860-raw-cel-1561690480.cel | 18 | 5 | HGPS | hgu133a | HGPS | AG11498 |