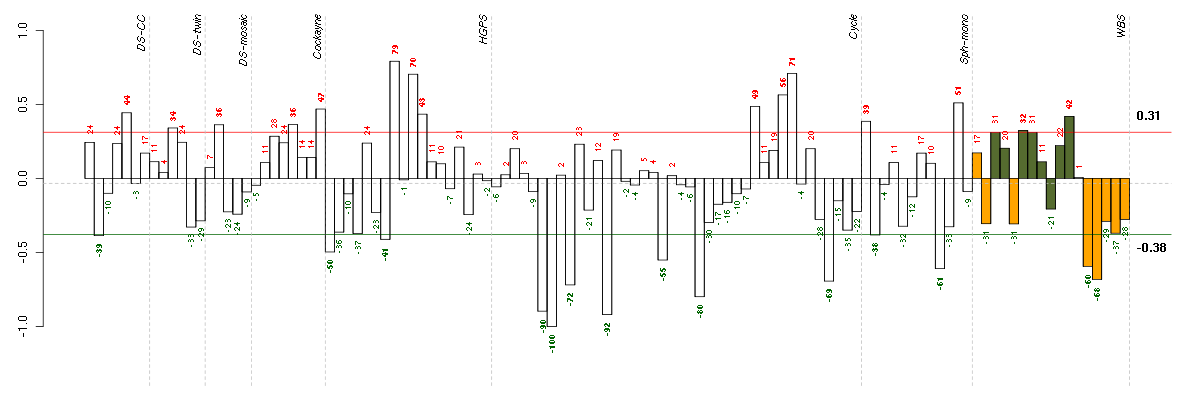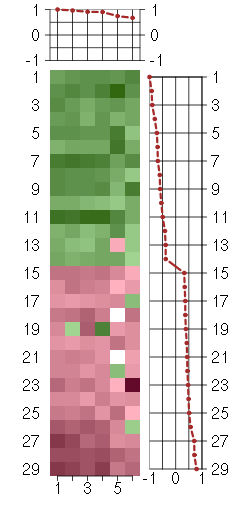

Under-expression is coded with green,
over-expression with red color.



CRTC3CREB regulated transcription coactivator 3 (218648_at), score: 0.74 DMWDdystrophia myotonica, WD repeat containing (213231_at), score: 0.67 LOC100132540similar to LOC339047 protein (214870_x_at), score: 0.96 LOC339047hypothetical protein LOC339047 (221501_x_at), score: 1 LOC399491LOC399491 protein (214035_x_at), score: 0.91 NPIPnuclear pore complex interacting protein (204538_x_at), score: 0.9
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| F055_WBS.CEL | 14 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| E-GEOD-4219-raw-cel-1311956457.cel | 14 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| D890_WBS.CEL | 13 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| E-TABM-263-raw-cel-1515486011.cel | 19 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690199.cel | 1 | 5 | HGPS | hgu133a | none | GM0316B |
| E-GEOD-3860-raw-cel-1561690272.cel | 7 | 5 | HGPS | hgu133a | HGPS | AG11498 |
| ctrl b 08-03.CEL | 2 | 1 | DS-CC | hgu133a | none | DS-CC 2 |
| E-GEOD-4219-raw-cel-1311956138.cel | 4 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| 5042_CNTL.CEL | 6 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| 3Twin.CEL | 3 | 2 | DS-twin | hgu133plus2 | Down | DS-twin 3 |
| 46B.CEL | 2 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 2 |
| E-GEOD-3407-raw-cel-1437949721.cel | 5 | 4 | Cockayne | hgu133a | CS | CSB |
| E-GEOD-4219-raw-cel-1311956083.cel | 2 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| 9118_CNTL.CEL | 11 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| E-GEOD-3860-raw-cel-1561690352.cel | 11 | 5 | HGPS | hgu133a | HGPS | AG11498 |
| t21b 08-03.CEL | 5 | 1 | DS-CC | hgu133a | Down | DS-CC 5 |
| E-GEOD-3407-raw-cel-1437949938.cel | 8 | 4 | Cockayne | hgu133a | none | CSB |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-4219-raw-cel-1311956634.cel | 19 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-TABM-263-raw-cel-1515486271.cel | 32 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690344.cel | 10 | 5 | HGPS | hgu133a | none | GM00038C |
| E-TABM-263-raw-cel-1515486291.cel | 33 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690304.cel | 8 | 5 | HGPS | hgu133a | none | GMO8398C |