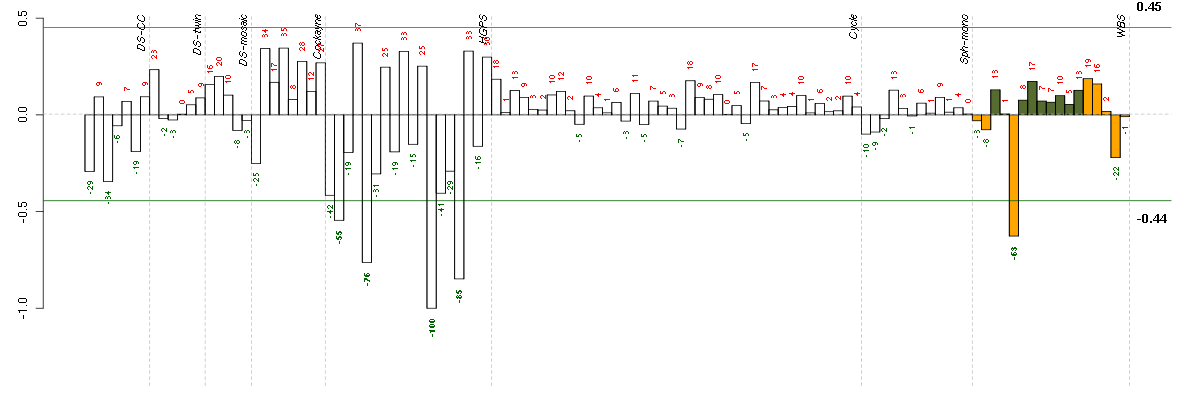

Under-expression is coded with green,
over-expression with red color.


extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
all
This term is the most general term possible

C20orf39chromosome 20 open reading frame 39 (219310_at), score: -0.72 C4orf31chromosome 4 open reading frame 31 (219747_at), score: -0.81 CGBchorionic gonadotropin, beta polypeptide (205387_s_at), score: -1 CKBcreatine kinase, brain (200884_at), score: -0.71 CLEC2BC-type lectin domain family 2, member B (209732_at), score: -0.72 COMPcartilage oligomeric matrix protein (205713_s_at), score: -0.77 EPHA4EPH receptor A4 (206114_at), score: -0.67 FAM46Afamily with sequence similarity 46, member A (221766_s_at), score: -0.71 GABRA5gamma-aminobutyric acid (GABA) A receptor, alpha 5 (206456_at), score: -0.89 HOXA11homeobox A11 (213823_at), score: -0.68 INHBBinhibin, beta B (205258_at), score: -0.71 MEGF6multiple EGF-like-domains 6 (213942_at), score: -0.75 NCAM1neural cell adhesion molecule 1 (212843_at), score: -0.71 NRG1neuregulin 1 (206343_s_at), score: -0.66 PLAC8placenta-specific 8 (219014_at), score: -0.9 PSG2pregnancy specific beta-1-glycoprotein 2 (208134_x_at), score: -0.72 PSG5pregnancy specific beta-1-glycoprotein 5 (204830_x_at), score: -0.69 PSG9pregnancy specific beta-1-glycoprotein 9 (209594_x_at), score: -0.68 PTPLBprotein tyrosine phosphatase-like (proline instead of catalytic arginine), member b (212640_at), score: -0.7 RUNX3runt-related transcription factor 3 (204198_s_at), score: -0.77 SALL1sal-like 1 (Drosophila) (206893_at), score: -0.68 SLC16A5solute carrier family 16, member 5 (monocarboxylic acid transporter 6) (206600_s_at), score: -0.7 STMN2stathmin-like 2 (203000_at), score: -0.67 UNC5Bunc-5 homolog B (C. elegans) (213100_at), score: -0.72
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-3860-raw-cel-1561690360.cel | 12 | 5 | HGPS | hgu133a | none | GM0316B |
| E-GEOD-3860-raw-cel-1561690416.cel | 15 | 5 | HGPS | hgu133a | none | GM0316B |
| E-GEOD-3860-raw-cel-1561690248.cel | 5 | 5 | HGPS | hgu133a | HGPS | AG11513 |
| 4319_WBS.CEL | 5 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| E-GEOD-3860-raw-cel-1561690215.cel | 2 | 5 | HGPS | hgu133a | HGPS | AG11513 |