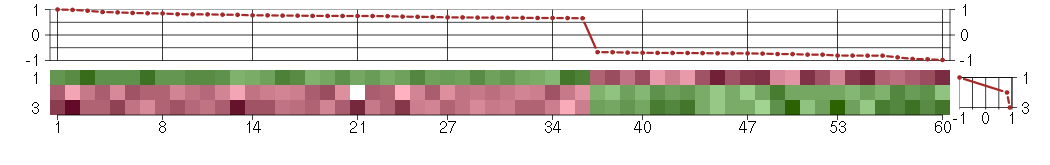

Under-expression is coded with green,
over-expression with red color.



protein binding
Interacting selectively with any protein or protein complex (a complex of two or more proteins that may include other nonprotein molecules).
molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
MAP-kinase scaffold activity
Functions as a physical support for the assembly of a multiprotein mitogen-activated protein kinase (MAPK) complex. MAPK scaffold proteins have binding sites for MAPK pathway kinases as well as for upstream signaling proteins.
structural molecule activity
The action of a molecule that contributes to the structural integrity of a complex or assembly within or outside a cell.
binding
The selective, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
receptor signaling complex scaffold activity
Functions to provide a physical support for the assembly of a multiprotein receptor signaling complex.
protein complex scaffold
Functions to provide a physical support for the assembly of a multiprotein complex.
all
This term is the most general term possible
protein complex scaffold
Functions to provide a physical support for the assembly of a multiprotein complex.
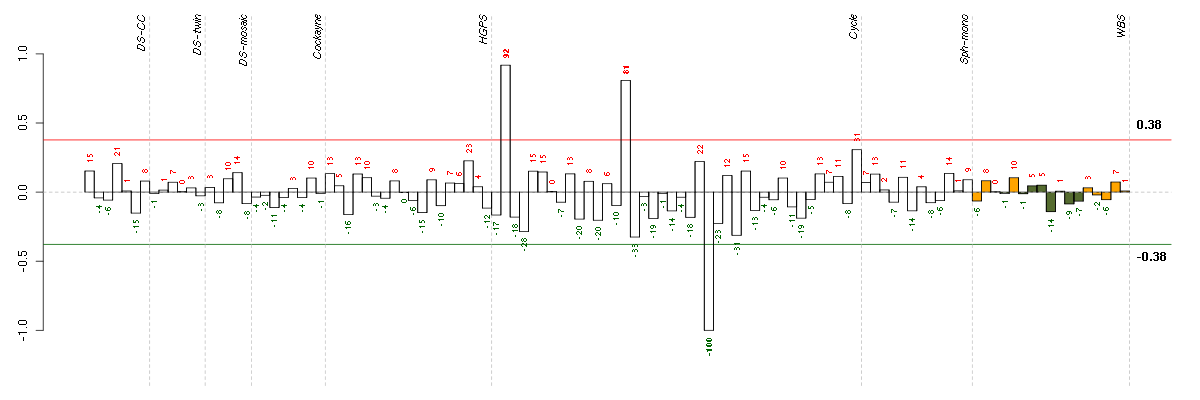
ABCA7ATP-binding cassette, sub-family A (ABC1), member 7 (219577_s_at), score: 0.98 ADORA1adenosine A1 receptor (216220_s_at), score: 0.68 ADRA1Dadrenergic, alpha-1D-, receptor (210961_s_at), score: 0.76 ANKRD46ankyrin repeat domain 46 (212731_at), score: -0.75 APPamyloid beta (A4) precursor protein (214953_s_at), score: 0.9 ASPHD1aspartate beta-hydroxylase domain containing 1 (214993_at), score: 0.75 BCAMbasal cell adhesion molecule (Lutheran blood group) (40093_at), score: 0.77 BCHEbutyrylcholinesterase (205433_at), score: -0.68 BRAFv-raf murine sarcoma viral oncogene homolog B1 (206044_s_at), score: -0.95 C18orf1chromosome 18 open reading frame 1 (209574_s_at), score: -0.73 CCDC40coiled-coil domain containing 40 (220592_at), score: 0.75 CNTLNcentlein, centrosomal protein (220095_at), score: -0.69 EDAectodysplasin A (211130_x_at), score: 0.85 EPS8L1EPS8-like 1 (218778_x_at), score: 0.82 EXPH5exophilin 5 (214734_at), score: -0.71 FKBP10FK506 binding protein 10, 65 kDa (219249_s_at), score: 1 GASTgastrin (208138_at), score: 0.81 GGT1gamma-glutamyltransferase 1 (209919_x_at), score: 0.68 GSDMBgasdermin B (215659_at), score: -0.77 HIPK2homeodomain interacting protein kinase 2 (219028_at), score: 0.67 HLA-DRB1major histocompatibility complex, class II, DR beta 1 (209312_x_at), score: 0.66 IL23Ainterleukin 23, alpha subunit p19 (211796_s_at), score: 0.72 INHAinhibin, alpha (210141_s_at), score: 0.75 ITGALintegrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide) (213475_s_at), score: 0.71 KLK13kallikrein-related peptidase 13 (205783_at), score: -0.81 KMOkynurenine 3-monooxygenase (kynurenine 3-hydroxylase) (205307_s_at), score: -0.72 KRT8keratin 8 (209008_x_at), score: 0.69 KRT8P12keratin 8 pseudogene 12 (222060_at), score: 0.79 LILRA5leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 5 (215838_at), score: 0.87 LOC100128919similar to HSPC157 (219865_at), score: -0.81 LRBALPS-responsive vesicle trafficking, beach and anchor containing (214109_at), score: -0.77 MAPK8IP1mitogen-activated protein kinase 8 interacting protein 1 (213013_at), score: 0.81 MAPK8IP2mitogen-activated protein kinase 8 interacting protein 2 (205050_s_at), score: 0.68 MDM2Mdm2 p53 binding protein homolog (mouse) (205386_s_at), score: -0.71 MGC13053hypothetical MGC13053 (211775_x_at), score: -0.81 MUC5ACmucin 5AC, oligomeric mucus/gel-forming (217182_at), score: 0.75 OPRL1opiate receptor-like 1 (206564_at), score: 0.69 PAX4paired box 4 (207867_at), score: -0.72 PEX13peroxisomal biogenesis factor 13 (205246_at), score: -0.67 PHC1polyhomeotic homolog 1 (Drosophila) (218338_at), score: -0.7 PHF2PHD finger protein 2 (212726_at), score: -0.75 PKLRpyruvate kinase, liver and RBC (222078_at), score: 0.74 PMS2L2postmeiotic segregation increased 2-like 2 pseudogene (215410_at), score: 0.8 POM121L2POM121 membrane glycoprotein-like 2 (rat) (216582_at), score: 0.95 RBM12BRNA binding motif protein 12B (51228_at), score: -0.88 RBM38RNA binding motif protein 38 (212430_at), score: 0.77 RLN1relaxin 1 (211753_s_at), score: 0.67 RNF121ring finger protein 121 (219021_at), score: 0.76 SCLYselenocysteine lyase (221575_at), score: -0.98 SEPT5septin 5 (209767_s_at), score: 0.71 SHANK2SH3 and multiple ankyrin repeat domains 2 (213307_at), score: -0.72 SLC30A4solute carrier family 30 (zinc transporter), member 4 (207362_at), score: -0.94 SLC5A5solute carrier family 5 (sodium iodide symporter), member 5 (211123_at), score: 0.66 ST8SIA3ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 (208064_s_at), score: 0.74 STXBP2syntaxin binding protein 2 (209367_at), score: 0.67 SUV39H2suppressor of variegation 3-9 homolog 2 (Drosophila) (219262_at), score: -0.71 TAF15TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa (202840_at), score: 0.85 TMPRSS6transmembrane protease, serine 6 (214955_at), score: 0.89 ZNF214zinc finger protein 214 (220497_at), score: -0.81 ZXDAzinc finger, X-linked, duplicated A (215263_at), score: -0.7
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486111.cel | 24 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485931.cel | 15 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |