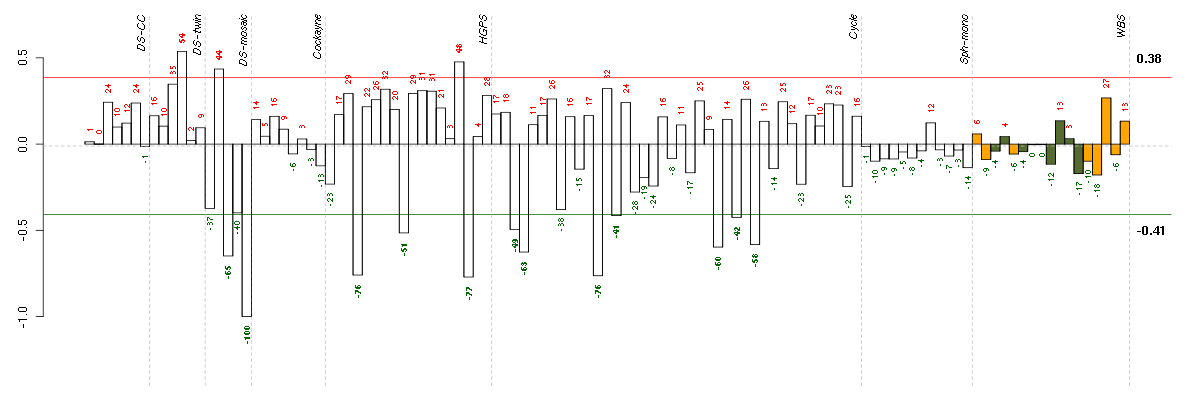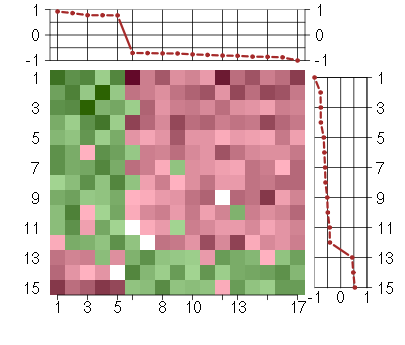

Under-expression is coded with green,
over-expression with red color.



ARHGDIBRho GDP dissociation inhibitor (GDI) beta (201288_at), score: -0.76 CYTIPcytohesin 1 interacting protein (209606_at), score: -0.71 ELK1ELK1, member of ETS oncogene family (210376_x_at), score: -0.71 FOXC2forkhead box C2 (MFH-1, mesenchyme forkhead 1) (214520_at), score: -0.88 GOLGA8Ggolgi autoantigen, golgin subfamily a, 8G (222149_x_at), score: 0.86 GPX3glutathione peroxidase 3 (plasma) (214091_s_at), score: -0.82 INAinternexin neuronal intermediate filament protein, alpha (204465_s_at), score: -0.85 LOC391132similar to hCG2041276 (216177_at), score: 0.77 LOC440434hypothetical protein FLJ11822 (215090_x_at), score: -0.72 MACROD1MACRO domain containing 1 (219188_s_at), score: 0.92 NOS1nitric oxide synthase 1 (neuronal) (207309_at), score: 0.78 PRSS3protease, serine, 3 (207463_x_at), score: -0.86 SLC22A17solute carrier family 22, member 17 (218675_at), score: 0.77 SLC29A1solute carrier family 29 (nucleoside transporters), member 1 (201801_s_at), score: -1 TMEM35transmembrane protein 35 (219685_at), score: -0.72 TOM1L2target of myb1-like 2 (chicken) (214840_at), score: -0.81 TRIM34tripartite motif-containing 34 (221044_s_at), score: -0.78
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| 47C.CEL | 5 | 3 | DS-mosaic | hgu133plus2 | Down mosaic | DS-mosaic 5 |
| E-GEOD-3860-raw-cel-1561690432.cel | 16 | 5 | HGPS | hgu133a | HGPS | AG10750 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690231.cel | 4 | 5 | HGPS | hgu133a | HGPS | AG10750 |
| 46C.CEL | 3 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 3 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486131.cel | 25 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690336.cel | 9 | 5 | HGPS | hgu133a | HGPS | AG10750 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486171.cel | 27 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485911.cel | 14 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 46B.CEL | 2 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 2 |
| E-GEOD-3860-raw-cel-1561690416.cel | 15 | 5 | HGPS | hgu133a | none | GM0316B |
| 4Twin.CEL | 4 | 2 | DS-twin | hgu133plus2 | none | DS-twin 4 |