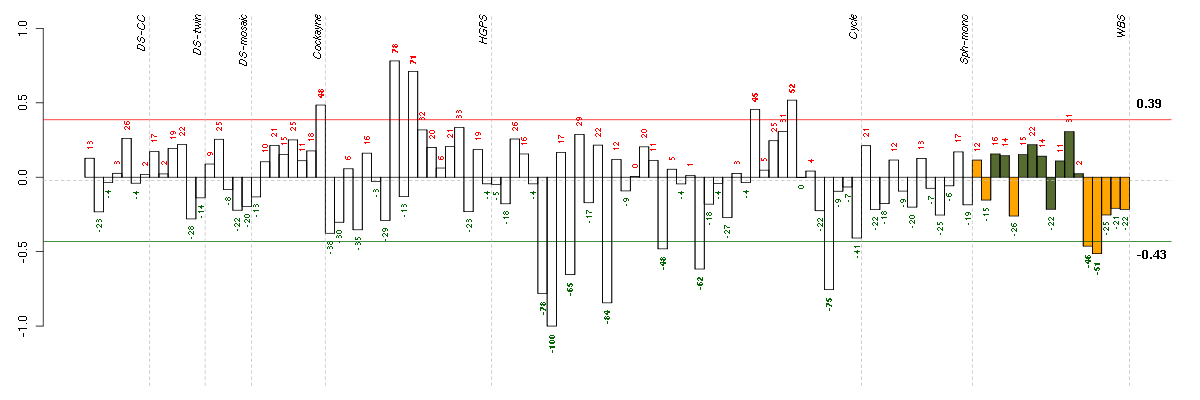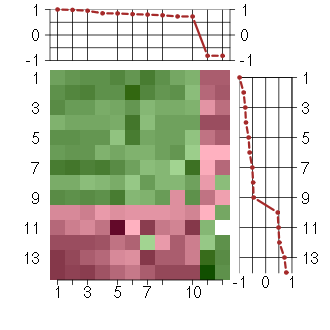

Under-expression is coded with green,
over-expression with red color.



CRTC3CREB regulated transcription coactivator 3 (218648_at), score: 0.82 DMWDdystrophia myotonica, WD repeat containing (213231_at), score: 0.85 FLJ12529pre-mRNA cleavage factor I, 59 kDa subunit (217866_at), score: 0.8 LOC100132540similar to LOC339047 protein (214870_x_at), score: 0.98 LOC339047hypothetical protein LOC339047 (221501_x_at), score: 1 LOC399491LOC399491 protein (214035_x_at), score: 0.85 LRRN3leucine rich repeat neuronal 3 (209841_s_at), score: -0.81 NEFMneurofilament, medium polypeptide (205113_at), score: -0.82 NPIPnuclear pore complex interacting protein (204538_x_at), score: 0.95 PKD1polycystic kidney disease 1 (autosomal dominant) (202328_s_at), score: 0.73 RNF220ring finger protein 220 (219988_s_at), score: 0.73 SCAMP4secretory carrier membrane protein 4 (213244_at), score: 0.77
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| F055_WBS.CEL | 14 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| E-TABM-263-raw-cel-1515486011.cel | 19 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| D890_WBS.CEL | 13 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3407-raw-cel-1437949938.cel | 8 | 4 | Cockayne | hgu133a | none | CSB |
| E-TABM-263-raw-cel-1515486291.cel | 33 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690344.cel | 10 | 5 | HGPS | hgu133a | none | GM00038C |
| E-GEOD-3860-raw-cel-1561690304.cel | 8 | 5 | HGPS | hgu133a | none | GMO8398C |