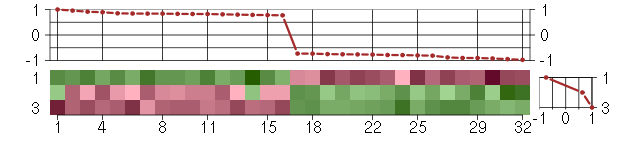

Under-expression is coded with green,
over-expression with red color.



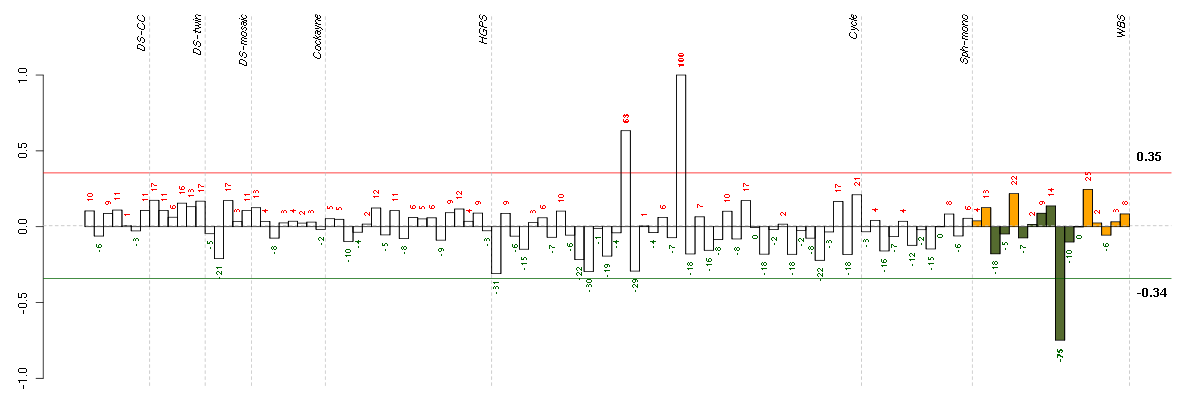
ACTR8ARP8 actin-related protein 8 homolog (yeast) (218658_s_at), score: -0.97 B3GALT4UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 4 (210205_at), score: 0.84 BCL2L11BCL2-like 11 (apoptosis facilitator) (222343_at), score: 0.82 C10orf28chromosome 10 open reading frame 28 (210455_at), score: -0.9 CFHR2complement factor H-related 2 (206910_x_at), score: 0.82 CHST2carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2 (203921_at), score: -0.76 CLIC2chloride intracellular channel 2 (213415_at), score: -0.88 CPZcarboxypeptidase Z (211062_s_at), score: -0.8 CYorf14chromosome Y open reading frame 14 (207063_at), score: -0.9 DDX28DEAD (Asp-Glu-Ala-Asp) box polypeptide 28 (40255_at), score: 0.84 HTR2B5-hydroxytryptamine (serotonin) receptor 2B (206638_at), score: 0.77 IL15interleukin 15 (205992_s_at), score: -0.78 ITGALintegrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide) (213475_s_at), score: 0.89 KIAA0644KIAA0644 gene product (205150_s_at), score: -0.76 LBA1lupus brain antigen 1 (213261_at), score: -0.77 LHX3LIM homeobox 3 (221670_s_at), score: 1 LOC388152hypothetical LOC388152 (220602_s_at), score: 0.78 LOC65998hypothetical protein LOC65998 (218641_at), score: -0.81 MALmal, T-cell differentiation protein (204777_s_at), score: 0.84 NFE2L3nuclear factor (erythroid-derived 2)-like 3 (204702_s_at), score: 0.79 NLKnemo-like kinase (218318_s_at), score: -0.73 PAFAH2platelet-activating factor acetylhydrolase 2, 40kDa (205232_s_at), score: 0.83 PGLYRP4peptidoglycan recognition protein 4 (220944_at), score: 0.96 PIN1Lpeptidylprolyl cis/trans isomerase, NIMA-interacting 1-like (pseudogene) (207582_at), score: 0.91 PIPOXpipecolic acid oxidase (221605_s_at), score: 0.81 PSTPIP2proline-serine-threonine phosphatase interacting protein 2 (219938_s_at), score: -0.73 RNASELribonuclease L (2',5'-oligoisoadenylate synthetase-dependent) (221287_at), score: -0.75 SASH3SAM and SH3 domain containing 3 (204923_at), score: 0.85 STIM1stromal interaction molecule 1 (202764_at), score: -0.95 TRAF1TNF receptor-associated factor 1 (205599_at), score: 0.8 TRPC6transient receptor potential cation channel, subfamily C, member 6 (217287_s_at), score: -0.92 ZNF675zinc finger protein 675 (217547_x_at), score: -0.79
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| 8495_CNTL.CEL | 10 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| E-TABM-263-raw-cel-1515485931.cel | 15 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486051.cel | 21 | 6 | Cycle | hgu133a2 | none | Cycle 1 |